 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pahal.C03047.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 846aa MW: 92962.7 Da PI: 6.2639 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 111.8 | 4e-35 | 164 | 238 | 2 | 76 |
-SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S- CS
SBP 2 CqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkk 76
Cqv+gCead+ e+k yhrrh+vC +++a++v+++g+++r+CqqC++fh l +fDe+krsCrr+L++hn+rrr+k
Pahal.C03047.1 164 CQVPGCEADIRELKGYHRRHRVCLRCAHASAVMLDGVQKRYCQQCGKFHVLLDFDEDKRSCRRKLERHNKRRRRK 238
************************************************************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51141 | 29.556 | 161 | 238 | IPR004333 | Transcription factor, SBP-box |
| Gene3D | G3DSA:4.10.1100.10 | 1.4E-26 | 161 | 225 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 2.09E-33 | 162 | 241 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 5.8E-27 | 164 | 238 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0055070 | Biological Process | copper ion homeostasis | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 846 aa Download sequence Send to blast |
DASDSGGGAS ADAGEQLWDW GNLLNFVQDD DSLVLPWDSD VGIGAADPTE AVAPLLPAPL 60 PQPEPEPGPV LPPPPLRVQG IGRRVRKRDP RLVCPNYLAG RVPCACPEVD EMVAAAEVED 120 VATEFLAGAR KKTKTAARRG KAGAAGGAGA AGGTVRAAAM EMKCQVPGCE ADIRELKGYH 180 RRHRVCLRCA HASAVMLDGV QKRYCQQCGK FHVLLDFDED KRSCRRKLER HNKRRRRKPD 240 SKGILDKEID EQLDLSADVS GDGELREENM EGTTSEMLET VLSNKILDRG TPAGSEDVLS 300 SPTCTQPSLQ NEQSKSVVTF AASVEACIGA KQENAKLTTN SPMHDTKSAY SSSCPTGRIS 360 FKLYDWNPAE FPRRLRHQIF EWLASMPVEL EGYIRPGCTI LTVFVAMPQH MWDKLSDDAA 420 NLLRNLVNSP NSLLLGKGAF FIHVNNMIFQ VLKDGATLMS TRLDVQSPRI DYVHPTWFEA 480 GKPVELILCG SSLDQPKFRS LLSFDGDYLK HDCHRLTSCD AIDCVENGDL ILDSQHEIFR 540 INITQSRPDI HGPAFVEVEN MFGLSNFVPI LFGSKQLCSE LERIQDALCG SSNSNLFGEL 600 PGAASDPYEH RKLQMAAMSG FLIDIGWLIR KPTHDDFKNV LSSTNIQRWV CILKFLIQND 660 FLNVLEIIVK SMENIMGSEV LSNLERGRLE DHVTAFLGYV SHARNIINSR ANSDEKTKLE 720 TRWISVSSPN QPSLGTSDPP ANETTGTGGD KNLHSANAAY EEETVPLVTR DDSHRHWCQP 780 DMNARWLKPS LVVKYPGGAT RMRLGMTVVV AAVLCFTACL VLFHPHGVGA LASPVKRYLS 840 SDSAL* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul5_A | 1e-37 | 163 | 243 | 5 | 85 | squamosa promoter binding protein-like 7 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 220 | 238 | KRSCRRKLERHNKRRRRKP |
| 2 | 224 | 236 | RRKLERHNKRRRR |
| 3 | 231 | 238 | NKRRRRKP |
| 4 | 232 | 237 | KRRRRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00604 | PBM | Transfer from AT5G18830 | Download |
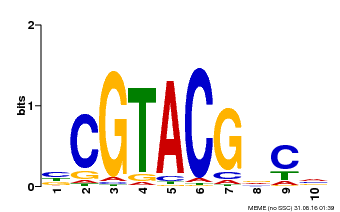
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025805456.1 | 0.0 | squamosa promoter-binding-like protein 9 | ||||
| Swissprot | Q6I576 | 0.0 | SPL9_ORYSJ; Squamosa promoter-binding-like protein 9 | ||||
| TrEMBL | A0A2T8KJS3 | 0.0 | A0A2T8KJS3_9POAL; Uncharacterized protein | ||||
| STRING | GRMZM2G109354_P01 | 0.0 | (Zea mays) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6922 | 33 | 44 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G18830.3 | 1e-138 | squamosa promoter binding protein-like 7 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pahal.C03047.1 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



