 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pahal.C03047.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 845aa MW: 92847.6 Da PI: 6.3341 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 111.8 | 4e-35 | 163 | 237 | 2 | 76 |
-SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S- CS
SBP 2 CqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkk 76
Cqv+gCead+ e+k yhrrh+vC +++a++v+++g+++r+CqqC++fh l +fDe+krsCrr+L++hn+rrr+k
Pahal.C03047.2 163 CQVPGCEADIRELKGYHRRHRVCLRCAHASAVMLDGVQKRYCQQCGKFHVLLDFDEDKRSCRRKLERHNKRRRRK 237
************************************************************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 1.4E-26 | 160 | 224 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 29.556 | 160 | 237 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 2.09E-33 | 161 | 240 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 5.8E-27 | 163 | 237 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0055070 | Biological Process | copper ion homeostasis | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 845 aa Download sequence Send to blast |
ASDSGGGASA DAGEQLWDWG NLLNFVQDDD SLVLPWDSDV GIGAADPTEA VAPLLPAPLP 60 QPEPEPGPVL PPPPLRVQGI GRRVRKRDPR LVCPNYLAGR VPCACPEVDE MVAAAEVEDV 120 ATEFLAGARK KTKTAARRGK AGAAGGAGAA GGTVRAAAME MKCQVPGCEA DIRELKGYHR 180 RHRVCLRCAH ASAVMLDGVQ KRYCQQCGKF HVLLDFDEDK RSCRRKLERH NKRRRRKPDS 240 KGILDKEIDE QLDLSADVSG DGELREENME GTTSEMLETV LSNKILDRGT PAGSEDVLSS 300 PTCTQPSLQN EQSKSVVTFA ASVEACIGAK QENAKLTTNS PMHDTKSAYS SSCPTGRISF 360 KLYDWNPAEF PRRLRHQIFE WLASMPVELE GYIRPGCTIL TVFVAMPQHM WDKLSDDAAN 420 LLRNLVNSPN SLLLGKGAFF IHVNNMIFQV LKDGATLMST RLDVQSPRID YVHPTWFEAG 480 KPVELILCGS SLDQPKFRSL LSFDGDYLKH DCHRLTSCDA IDCVENGDLI LDSQHEIFRI 540 NITQSRPDIH GPAFVEVENM FGLSNFVPIL FGSKQLCSEL ERIQDALCGS SNSNLFGELP 600 GAASDPYEHR KLQMAAMSGF LIDIGWLIRK PTHDDFKNVL SSTNIQRWVC ILKFLIQNDF 660 LNVLEIIVKS MENIMGSEVL SNLERGRLED HVTAFLGYVS HARNIINSRA NSDEKTKLET 720 RWISVSSPNQ PSLGTSDPPA NETTGTGGDK NLHSANAAYE EETVPLVTRD DSHRHWCQPD 780 MNARWLKPSL VVKYPGGATR MRLGMTVVVA AVLCFTACLV LFHPHGVGAL ASPVKRYLSS 840 DSAL* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul5_A | 1e-37 | 162 | 242 | 5 | 85 | squamosa promoter binding protein-like 7 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 219 | 237 | KRSCRRKLERHNKRRRRKP |
| 2 | 223 | 235 | RRKLERHNKRRRR |
| 3 | 230 | 237 | NKRRRRKP |
| 4 | 231 | 236 | KRRRRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
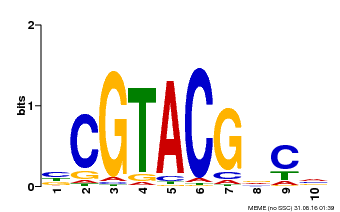
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00604 | PBM | Transfer from AT5G18830 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025805456.1 | 0.0 | squamosa promoter-binding-like protein 9 | ||||
| Swissprot | Q6I576 | 0.0 | SPL9_ORYSJ; Squamosa promoter-binding-like protein 9 | ||||
| TrEMBL | A0A2T8KJS3 | 0.0 | A0A2T8KJS3_9POAL; Uncharacterized protein | ||||
| STRING | GRMZM2G109354_P01 | 0.0 | (Zea mays) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G18830.3 | 1e-138 | squamosa promoter binding protein-like 7 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pahal.C03047.2 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



