 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pahal.C03047.3 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 642aa MW: 71791.6 Da PI: 6.3147 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 36 | 1.9e-11 | 6 | 34 | 48 | 76 |
SEEETTT--SS--S-STTTT-------S- CS
SBP 48 rfhelsefDeekrsCrrrLakhnerrrkk 76
rfh l +fDe+krsCrr+L++hn+rrr+k
Pahal.C03047.3 6 RFHVLLDFDEDKRSCRRKLERHNKRRRRK 34
9*************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51141 | 12.921 | 1 | 34 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 1.5E-5 | 6 | 34 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 3.14E-9 | 6 | 37 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0055070 | Biological Process | copper ion homeostasis | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 642 aa Download sequence Send to blast |
MYFHFRFHVL LDFDEDKRSC RRKLERHNKR RRRKPDSKGI LDKEIDEQLD LSADVSGDGE 60 LREENMEGTT SEMLETVLSN KILDRGTPAG SEDVLSSPTC TQPSLQNEQS KSVVTFAASV 120 EACIGAKQEN AKLTTNSPMH DTKSAYSSSC PTGRISFKLY DWNPAEFPRR LRHQIFEWLA 180 SMPVELEGYI RPGCTILTVF VAMPQHMWDK LSDDAANLLR NLVNSPNSLL LGKGAFFIHV 240 NNMIFQVLKD GATLMSTRLD VQSPRIDYVH PTWFEAGKPV ELILCGSSLD QPKFRSLLSF 300 DGDYLKHDCH RLTSCDAIDC VENGDLILDS QHEIFRINIT QSRPDIHGPA FVEVENMFGL 360 SNFVPILFGS KQLCSELERI QDALCGSSNS NLFGELPGAA SDPYEHRKLQ MAAMSGFLID 420 IGWLIRKPTH DDFKNVLSST NIQRWVCILK FLIQNDFLNV LEIIVKSMEN IMGSEVLSNL 480 ERGRLEDHVT AFLGYVSHAR NIINSRANSD EKTKLETRWI SVSSPNQPSL GTSDPPANET 540 TGTGGDKNLH SANAAYEEET VPLVTRDDSH RHWCQPDMNA RWLKPSLVVK YPGGATRMRL 600 GMTVVVAAVL CFTACLVLFH PHGVGALASP VKRYLSSDSA L* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 16 | 34 | KRSCRRKLERHNKRRRRKP |
| 2 | 20 | 32 | RRKLERHNKRRRR |
| 3 | 27 | 34 | NKRRRRKP |
| 4 | 28 | 33 | KRRRRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
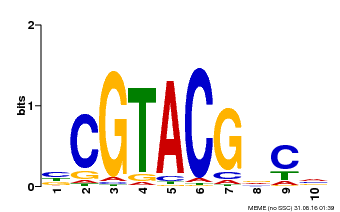
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00604 | PBM | Transfer from AT5G18830 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025805456.1 | 0.0 | squamosa promoter-binding-like protein 9 | ||||
| Swissprot | Q6I576 | 0.0 | SPL9_ORYSJ; Squamosa promoter-binding-like protein 9 | ||||
| TrEMBL | A0A2T8KJS3 | 0.0 | A0A2T8KJS3_9POAL; Uncharacterized protein | ||||
| STRING | Sb09g020110.1 | 0.0 | (Sorghum bicolor) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G18830.3 | 1e-109 | squamosa promoter binding protein-like 7 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pahal.C03047.3 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



