 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pahal.C03772.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 279aa MW: 29742.3 Da PI: 9.6155 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 145.8 | 2.3e-45 | 14 | 151 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdL.pkkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevls 94
lppGfrF+Ptdeelvv+yL++++ +++l+++ i++v+i+ ++P+dL p +e+e yfF++++ ky +g+r+nrat +gyWkatgk+k+v++
Pahal.C03772.1 14 LPPGFRFRPTDEELVVHYLRRRALASPLPAAVDIPDVRILAHDPSDLlP--PGWSEQERYFFTCKEAKYVKGRRANRATGAGYWKATGKEKPVVV 106
79***************************9666**************43..3447899************************************9 PP
NAM 95 k...........kgelvglkktLvfykgrapkgektdWvmheyrl 128
+ + lvg+k++Lvfy+g+ p+g+ktdWvmheyrl
Pahal.C03772.1 107 AvparaggqgkaQAVLVGMKRSLVFYRGKPPTGSKTDWVMHEYRL 151
9999887765444444***************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 1.7E-49 | 9 | 185 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 48.314 | 14 | 185 | IPR003441 | NAC domain |
| Pfam | PF02365 | 2.0E-26 | 15 | 151 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0010089 | Biological Process | xylem development | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 279 aa Download sequence Send to blast |
MEAAKEPAVT KPRLPPGFRF RPTDEELVVH YLRRRALASP LPAAVDIPDV RILAHDPSDL 60 LPPGWSEQER YFFTCKEAKY VKGRRANRAT GAGYWKATGK EKPVVVAVPA RAGGQGKAQA 120 VLVGMKRSLV FYRGKPPTGS KTDWVMHEYR LAGAGLAPCR RAAGSQAAEA SRPAEGWVLC 180 RVFRKKGSAS ANAAAAAAAD AEASPADGGS DGEAEDAGEE EGGRTFIDFF ARADADAAGR 240 RLQQQERRAS SPVVSSSCLT DASHEQHGRE QETTSRGA* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 4e-38 | 9 | 189 | 12 | 169 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 4e-38 | 9 | 189 | 12 | 169 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 4e-38 | 9 | 189 | 12 | 169 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 4e-38 | 9 | 189 | 12 | 169 | NO APICAL MERISTEM PROTEIN |
| 4dul_A | 4e-38 | 9 | 189 | 12 | 169 | NAC domain-containing protein 19 |
| 4dul_B | 4e-38 | 9 | 189 | 12 | 169 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional repressor that negatively regulates the expression of genes involved in xylem vessel formation. Represses the transcriptional activation activity of NAC030/VND7, which regulates protoxylem vessel differentiation by promoting immature xylem vessel-specific genes expression (PubMed:20388856). Transcriptional activator that regulates the COLD-REGULATED (COR15A and COR15B) and RESPONSIVE TO DEHYDRATION (LTI78/RD29A and LTI65/RD29B) genes by binding directly to their promoters. Mediates signaling crosstalk between salt stress response and leaf aging process (PubMed:21673078). May play a role in DNA replication of mungbean yellow mosaic virus (PubMed:24442717). {ECO:0000269|PubMed:20388856, ECO:0000269|PubMed:21673078, ECO:0000269|PubMed:24442717}. | |||||
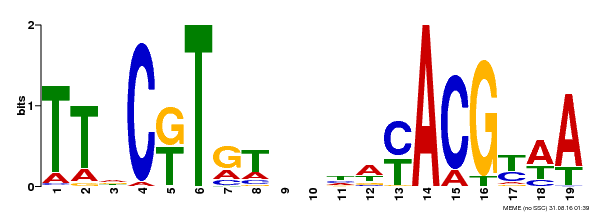
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00507 | DAP | Transfer from AT5G13180 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By abscisic acid (ABA) and salt stress. {ECO:0000269|PubMed:21673078}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025809222.1 | 0.0 | NAC domain-containing protein 83 | ||||
| Swissprot | Q9FY93 | 7e-56 | NAC83_ARATH; NAC domain-containing protein 83 | ||||
| TrEMBL | A0A2S3HE73 | 0.0 | A0A2S3HE73_9POAL; Uncharacterized protein | ||||
| STRING | Pavir.Cb00769.1.p | 1e-137 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6317 | 32 | 54 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G13180.1 | 1e-46 | NAC domain containing protein 83 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pahal.C03772.1 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



