 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pahal.D02122.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 737aa MW: 77470.4 Da PI: 6.0716 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 100.3 | 1.6e-31 | 549 | 633 | 1 | 86 |
trihelix 1 rWtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdqle 86
rW+k+ev aLi++r+ +e r+++ lk+plWeevs++m++ g+ rs+k+Ckekwen+nk+++k ke+ kkr + + +tcpyfd+l+
Pahal.D02122.1 549 RWPKHEVEALIRVRTGLEGRFQEPGLKGPLWEEVSARMAAAGYGRSAKRCKEKWENINKYFRKAKESGKKR-PAHAKTCPYFDELD 633
8********************************************************************98.99999*******98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50090 | 7.317 | 542 | 606 | IPR017877 | Myb-like domain |
| Gene3D | G3DSA:1.10.10.60 | 6.2E-4 | 545 | 605 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 0.0065 | 546 | 608 | IPR001005 | SANT/Myb domain |
| Pfam | PF13837 | 7.9E-23 | 548 | 635 | No hit | No description |
| CDD | cd12203 | 9.41E-30 | 548 | 613 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 737 aa Download sequence Send to blast |
MQSGYGGVSE FQQFIMDGGF AMSAPPQQPQ PPQPAAAQEL GGPFRYQPLH HHALPPQHHH 60 HAPHMPPHFA HFGAPPQAPF TQQLLHQAAA AGHHHHLQLF HEQHHHHKPP PPQPQQHQPQ 120 QSAPSRWAPQ HHHQQPHQQH HHPHHLGFDV EAVPESSGAG AGSAASGGAA PPGVPPFLAA 180 AMNFKLAVDA GGGSGATGGT DDALNDGGGG AGSGMMLHVG GGGGGGDDEA APESRLRRWN 240 GDEETSIKEP TWRPLDIDYL HSTSSSKRAP GKEKVATPES PAPTAAAANY FKKGDDNAAA 300 AAAAAAAASA GGSNYKLFSE LEAIYKPGSS GAGGAQTGSG SGLTGDDNAI LEPAMADLPG 360 VAAADAPQLN TSETSAGEDA AAVVQPPQPQ PSADAARRKR KRRRQEQLSA SASFFERLVQ 420 RLMEHQESLH RQFLDAMERR ERERAARDEA WRRQEADKFA REATARAQDR ASAAAREAAI 480 IAYLEKISGE SIALPPPAAA SGDDMSQDAA AGKELVPYDG GGGGGGGVCG DTAPGSGGDV 540 GSLHLSTSRW PKHEVEALIR VRTGLEGRFQ EPGLKGPLWE EVSARMAAAG YGRSAKRCKE 600 KWENINKYFR KAKESGKKRP AHAKTCPYFD ELDRLYSRTG GQSNNGSAGS NSNAGGGDEA 660 KASSELLDAV VRYPDAQGGP PGFMFDREHQ NEAGGREEGA ADEEEDGGIA KGRAADDEDQ 720 DQDDEVESHG GHDDDE* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 396 | 401 | RRKRKR |
| 2 | 397 | 402 | RKRKRR |
| 3 | 397 | 403 | RKRKRRR |
| 4 | 398 | 403 | KRKRRR |
| 5 | 399 | 403 | RKRRR |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00651 | PBM | Transfer from LOC_Os02g01380 | Download |
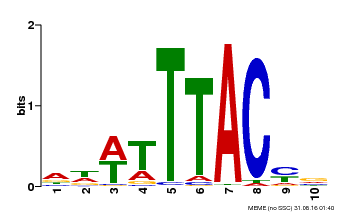
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT088333 | 1e-180 | BT088333.2 Zea mays full-length cDNA clone ZM_BFc0144A13 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025819158.1 | 0.0 | trihelix transcription factor GT-2-like | ||||
| TrEMBL | A0A2S3GKE4 | 0.0 | A0A2S3GKE4_9POAL; Uncharacterized protein | ||||
| STRING | Sb04g000520.1 | 0.0 | (Sorghum bicolor) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6399 | 35 | 47 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G76890.2 | 9e-39 | Trihelix family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pahal.D02122.1 |



