 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pahal.E04294.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 293aa MW: 30647.4 Da PI: 10.6338 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 48.3 | 2.3e-15 | 118 | 162 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT++E+ +++ + ++lG+g+W+ I+r++ ++Rt+ q+ s+ qky
Pahal.E04294.1 118 PWTEDEHRRFLAGLEKLGKGDWRGISRHFVTTRTPTQVASHAQKY 162
8*******************************************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50158 | 8.581 | 3 | 18 | IPR001878 | Zinc finger, CCHC-type |
| PROSITE profile | PS51294 | 20.171 | 111 | 167 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.92E-18 | 112 | 168 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 8.1E-19 | 114 | 166 | IPR006447 | Myb domain, plants |
| Gene3D | G3DSA:1.10.10.60 | 2.1E-11 | 115 | 161 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.5E-11 | 115 | 165 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.3E-12 | 118 | 162 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 5.34E-10 | 118 | 163 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 293 aa Download sequence Send to blast |
MARKCSSCGH NGHNSRTCSG HGRVSETGGG GGGVRLFGVQ LHVGPSSSPM KKCFSMDCLS 60 SAAPPAYYAA ALAASSSSPS VSSSSSLVSV EETAEKVNNG YLSDGLMGRA QERKKGVPWT 120 EDEHRRFLAG LEKLGKGDWR GISRHFVTTR TPTQVASHAQ KYFLRQSSLT QKKRRSSLFD 180 VVENAGRAAS SAPDAELPAL SLGISRPAAP SRVLPPTLSS LLPQCSAPTS RSGSGSTSPS 240 GIAAPKHRHP PSSLTPVSKT HQAPDLELKI STADHQTGSS PRTPFFGTIR VT* |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00565 | DAP | Transfer from AT5G56840 | Download |
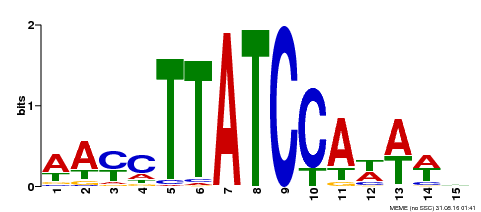
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EU967099 | 1e-153 | EU967099.1 Zea mays clone 299455 mRNA sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025818314.1 | 0.0 | transcription factor MYBS3-like | ||||
| TrEMBL | A0A2S3HYJ5 | 0.0 | A0A2S3HYJ5_9POAL; Uncharacterized protein | ||||
| STRING | Pavir.Ea00361.1.p | 1e-142 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP4231 | 38 | 73 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G56840.1 | 1e-47 | MYB_related family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pahal.E04294.1 |



