 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pahal.I00587.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 281aa MW: 30558.2 Da PI: 6.8655 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 56.1 | 6.2e-18 | 61 | 114 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k+++++ eq+++Le+ Fe +++ e++ +LA+ lgL+ rqV +WFqNrRa++k
Pahal.I00587.2 61 KKRRLNVEQVRTLEKNFELGNKLEPERKLQLARALGLQPRQVAIWFQNRRARWK 114
4568999**********************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 128.4 | 2.9e-41 | 60 | 150 | 1 | 91 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreel 91
ekkrrl+ eqv++LE++Fe +kLeperK +lar+Lglqprqva+WFqnrRAR+ktkqlEkdy+aLkr++da+k++n++L +++++L++e+
Pahal.I00587.2 60 EKKRRLNVEQVRTLEKNFELGNKLEPERKLQLARALGLQPRQVAIWFQNRRARWKTKQLEKDYDALKRQLDAVKADNDALLSHNKKLQAEI 150
69**************************************************************************************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 4.9E-19 | 37 | 117 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 1.54E-19 | 52 | 118 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.346 | 56 | 116 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.3E-17 | 59 | 120 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 2.4E-15 | 61 | 114 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 3.87E-16 | 61 | 117 | No hit | No description |
| PRINTS | PR00031 | 1.2E-5 | 87 | 96 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 91 | 114 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 1.2E-5 | 96 | 112 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 4.8E-14 | 116 | 156 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009744 | Biological Process | response to sucrose | ||||
| GO:0048826 | Biological Process | cotyledon morphogenesis | ||||
| GO:0080022 | Biological Process | primary root development | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 281 aa Download sequence Send to blast |
MPLPPRLSRK LIHTARMDAG MAPMLGKRPM YGAEAGGGDE ANGGGGNEDE LSDDGSQAGE 60 KKRRLNVEQV RTLEKNFELG NKLEPERKLQ LARALGLQPR QVAIWFQNRR ARWKTKQLEK 120 DYDALKRQLD AVKADNDALL SHNKKLQAEI LALKGREAGS ELINLNKETE ASCSNRSENS 180 SEINLDISRT PPSEGPDPPP SHHQHPGGGG GGMIPFYPSV GGRPAGVDMD QLLHSTSGPK 240 LEQHGNGGVQ APETASFGNL LCGVDEPPPF WPWADHQHFH * |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 108 | 116 | RRARWKTKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
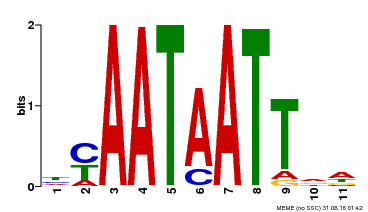
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00225 | DAP | Transfer from AT1G69780 | Download |
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EU966490 | 0.0 | EU966490.1 Zea mays clone 294849 homeobox-leucine zipper protein HAT7 mRNA, complete cds. | |||
| GenBank | KJ728506 | 0.0 | KJ728506.1 Zea mays clone pUT6811 HB transcription factor (HB102) mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025796010.1 | 0.0 | homeobox-leucine zipper protein HOX21-like isoform X2 | ||||
| Swissprot | Q8S7W9 | 1e-133 | HOX21_ORYSJ; Homeobox-leucine zipper protein HOX21 | ||||
| TrEMBL | A0A2S3ITX4 | 0.0 | A0A2S3ITX4_9POAL; Uncharacterized protein | ||||
| TrEMBL | A0A2T7CH06 | 0.0 | A0A2T7CH06_9POAL; Uncharacterized protein | ||||
| STRING | Pavir.Ia04323.1.p | 0.0 | (Panicum virgatum) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G69780.1 | 1e-75 | HD-ZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pahal.I00587.2 |



