 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pahal.I00986.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | YABBY | ||||||||
| Protein Properties | Length: 196aa MW: 21892.3 Da PI: 8.7686 | ||||||||
| Description | YABBY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | YABBY | 57.7 | 5.2e-18 | 1 | 46 | 1 | 46 |
YABBY 1 advfssseqvCyvqCnfCntilavsvPstslfkvvtvrCGhCtsll 46
+d++s+se++Cyv+C++Cnt+lav vP + l+ +vtv+CGhC +l
Pahal.I00986.1 1 MDLVSQSEHLCYVRCTYCNTVLAVGVPCKRLMDTVTVKCGHCNNLS 46
57899**************************************984 PP
| |||||||
| 2 | YABBY | 93.9 | 3.8e-29 | 86 | 147 | 101 | 162 |
YABBY 101 lsenedeevprvppvirPPekrqrvPsaynrfikeeiqrikasnPdishreafsaaaknWah 162
s ++e +pr+p v++PPek+ r Psaynrf++eeiqrika+ Pdi hreafs+aaknWa
Pahal.I00986.1 86 PSPTSTEASPRMPFVVKPPEKKHRLPSAYNRFMREEIQRIKAAKPDIPHREAFSMAAKNWAK 147
377789999****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04690 | 2.1E-46 | 6 | 148 | IPR006780 | YABBY protein |
| SuperFamily | SSF47095 | 1.57E-8 | 90 | 149 | IPR009071 | High mobility group box domain |
| Gene3D | G3DSA:1.10.30.10 | 1.9E-5 | 102 | 149 | IPR009071 | High mobility group box domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010254 | Biological Process | nectary development | ||||
| GO:0010582 | Biological Process | floral meristem determinacy | ||||
| GO:0048479 | Biological Process | style development | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 196 aa Download sequence Send to blast |
MDLVSQSEHL CYVRCTYCNT VLAVGVPCKR LMDTVTVKCG HCNNLSYLSP RPPMVQPLSP 60 TDHPLGPFQC QGPCNDCRRN QPMPLPSPTS TEASPRMPFV VKPPEKKHRL PSAYNRFMRE 120 EIQRIKAAKP DIPHREAFSM AAKNWAKCDP RCSTTVSTAT SNSAPEPRAV PAPQERAKEQ 180 VIESFDIFKQ IERSI* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Regulates carpel specification in flower development. Severe or intermediate mutation in DL causes complete or partial homeotic conversion of carpels into stamens without affecting the identities of other floral organs. Interacts antagonistically with class B genes and controls floral meristem determinacy. Regulates midrib formation in leaves probably by inducing cell proliferation in the central region of the leaf. {ECO:0000269|PubMed:14729915}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00220 | DAP | Transfer from AT1G69180 | Download |
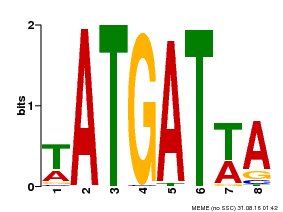
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AB470268 | 0.0 | AB470268.1 Sorghum bicolor SbDL mRNA for DL related protein, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025797474.1 | 1e-143 | protein DROOPING LEAF isoform X2 | ||||
| Swissprot | Q76EJ0 | 1e-116 | YABDL_ORYSJ; Protein DROOPING LEAF | ||||
| TrEMBL | A0A2S3IT10 | 1e-142 | A0A2S3IT10_9POAL; Uncharacterized protein | ||||
| TrEMBL | A0A2T7CFS6 | 1e-142 | A0A2T7CFS6_9POAL; Uncharacterized protein | ||||
| STRING | Pavir.Ib00755.1.p | 1e-140 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP5951 | 36 | 57 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G69180.1 | 4e-52 | YABBY family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pahal.I00986.1 |



