 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pahal.I01545.3 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | TALE | ||||||||
| Protein Properties | Length: 334aa MW: 36976.7 Da PI: 7.1192 | ||||||||
| Description | TALE family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 28.9 | 2e-09 | 250 | 285 | 20 | 55 |
HHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHH CS
Homeobox 20 eknrypsaeereeLAkklgLterqVkvWFqNrRake 55
+k +yps++e+ LA+ +gL+++q+ +WF N+R ++
Pahal.I01545.3 250 YKWPYPSETEKLALAETTGLDQKQINNWFINQRKRH 285
4679*****************************885 PP
| |||||||
| 2 | ELK | 43 | 9.3e-15 | 205 | 226 | 1 | 22 |
ELK 1 ELKhqLlrKYsgyLgsLkqEFs 22
ELKhqLlrKY+gyLg+L+qEFs
Pahal.I01545.3 205 ELKHQLLRKYGGYLGGLRQEFS 226
9********************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01255 | 2.7E-21 | 78 | 122 | IPR005540 | KNOX1 |
| Pfam | PF03790 | 7.6E-22 | 80 | 120 | IPR005540 | KNOX1 |
| SMART | SM01256 | 2.7E-29 | 130 | 181 | IPR005541 | KNOX2 |
| Pfam | PF03791 | 1.2E-23 | 135 | 180 | IPR005541 | KNOX2 |
| SMART | SM01188 | 4.2E-8 | 205 | 226 | IPR005539 | ELK domain |
| Pfam | PF03789 | 3.5E-11 | 205 | 226 | IPR005539 | ELK domain |
| PROSITE profile | PS51213 | 11.575 | 205 | 225 | IPR005539 | ELK domain |
| PROSITE profile | PS50071 | 12.989 | 225 | 288 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 1.5E-19 | 226 | 298 | IPR009057 | Homeodomain-like |
| SMART | SM00389 | 8.4E-12 | 227 | 292 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 3.0E-27 | 230 | 291 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 3.28E-12 | 237 | 288 | No hit | No description |
| Pfam | PF05920 | 2.0E-16 | 245 | 284 | IPR008422 | Homeobox KN domain |
| PROSITE pattern | PS00027 | 0 | 263 | 286 | IPR017970 | Homeobox, conserved site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 334 aa Download sequence Send to blast |
MDSFKDLGGG GSSASKASFL QLPLPASSSA QGFPSPDGHH HSSRLPLQQL LADPSGAQRN 60 HQMDGAVVQR EISPVDAEII KAKIMSHPQY SALVAAYLDC QKVGAPPDVS DRLSAMAAKL 120 DAQPGPSRRR REPARADPEL DQFMEAYCNM LVKYQEELAR PIQEAAEFFK SMERQLDSIT 180 GAGSSEDDQD GSCPEEIDPF AEDKELKHQL LRKYGGYLGG LRQEFSKRKK KGKLPKEARQ 240 KLLHWWELHY KWPYPSETEK LALAETTGLD QKQINNWFIN QRKRHWKPTS EDMPFAMMEA 300 AGGFHAPQGA AAALYVASRT PFMADGGMHR LGS* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may be involved in shoot formation during embryogenesis. {ECO:0000269|PubMed:10080693}. | |||||
| UniProt | Probable transcription factor that may be involved in shoot formation during embryogenesis. {ECO:0000269|PubMed:10080693, ECO:0000269|PubMed:9869405}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00671 | PBM | Transfer from GRMZM2G135447 | Download |
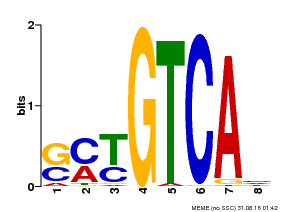
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025795147.1 | 0.0 | homeobox protein knotted-1-like 12 isoform X4 | ||||
| Swissprot | O65034 | 1e-115 | KNOSC_ORYSI; Homeobox protein knotted-1-like 12 | ||||
| Swissprot | O80416 | 1e-115 | KNOSC_ORYSJ; Homeobox protein knotted-1-like 12 | ||||
| TrEMBL | A0A2S3IIB4 | 0.0 | A0A2S3IIB4_9POAL; Uncharacterized protein | ||||
| STRING | Pavir.Ia00428.1.p | 0.0 | (Panicum virgatum) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G08150.1 | 2e-82 | KNOTTED-like from Arabidopsis thaliana | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pahal.I01545.3 |



