 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pahal.I01913.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | TALE | ||||||||
| Protein Properties | Length: 355aa MW: 39208.2 Da PI: 7.0487 | ||||||||
| Description | TALE family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 26.9 | 8e-09 | 282 | 317 | 20 | 55 |
HHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHH CS
Homeobox 20 eknrypsaeereeLAkklgLterqVkvWFqNrRake 55
+k +yps++++ LA+++gL+ +q+ +WF N+R ++
Pahal.I01913.1 282 YKWPYPSETQKVALAESTGLDLKQINNWFINQRKRH 317
5679*****************************885 PP
| |||||||
| 2 | ELK | 37.1 | 6.5e-13 | 237 | 258 | 1 | 22 |
ELK 1 ELKhqLlrKYsgyLgsLkqEFs 22
ELKh+Ll+KYsgyL+sLkqE+s
Pahal.I01913.1 237 ELKHHLLKKYSGYLSSLKQELS 258
9*******************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01255 | 2.3E-20 | 97 | 141 | IPR005540 | KNOX1 |
| Pfam | PF03790 | 3.1E-22 | 98 | 140 | IPR005540 | KNOX1 |
| SMART | SM01256 | 2.4E-27 | 148 | 199 | IPR005541 | KNOX2 |
| Pfam | PF03791 | 1.8E-22 | 154 | 197 | IPR005541 | KNOX2 |
| SMART | SM01188 | 570 | 157 | 177 | IPR005539 | ELK domain |
| Pfam | PF03789 | 8.0E-10 | 237 | 258 | IPR005539 | ELK domain |
| SMART | SM01188 | 4.9E-7 | 237 | 258 | IPR005539 | ELK domain |
| PROSITE profile | PS51213 | 11.331 | 237 | 257 | IPR005539 | ELK domain |
| PROSITE profile | PS50071 | 12.487 | 257 | 320 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 1.58E-19 | 258 | 333 | IPR009057 | Homeodomain-like |
| SMART | SM00389 | 4.2E-12 | 259 | 324 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 7.3E-28 | 262 | 322 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 8.41E-12 | 269 | 321 | No hit | No description |
| Pfam | PF05920 | 6.6E-16 | 277 | 316 | IPR008422 | Homeobox KN domain |
| PROSITE pattern | PS00027 | 0 | 295 | 318 | IPR017970 | Homeobox, conserved site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 355 aa Download sequence Send to blast |
MEEITHHFGV GASGHGHGQH HHHHHHPWGS SLSAVVAPPP QPPPRAGLPL TLNTAATVNS 60 GAGGNPVLQL ANGGSLLDAC IKAKEPSSSS LYAGDVEAIK AKIISHPHYY SLLAAYLECQ 120 KVGAPPEVSA RLTAMAQELE ARQRTALGGL GAATEPELDQ FMEAYHEMLV KFREELTRPL 180 QEAMEFMRRV ESQLSSLSIS GRSLRNILSS GSSEEDQEGS GGETELPEVD AHGVDQELKH 240 HLLKKYSGYL SSLKQELSKK KKKGKLPKEA RQQLLSWWDL HYKWPYPSET QKVALAESTG 300 LDLKQINNWF INQRKRHWKP SEEMHHLMMD GYHTTGAFYM DGHFINDGGL YRLG* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds to RNA (PubMed:17965274). Possible transcription factor that regulates genes involved in development. Mutations in KN-1 alter leaf development. Foci of cells along the lateral vein do not differentiate properly but continue to divide, forming knots. May participate in the switch from indeterminate to determinate cell fates. Probably binds to the DNA sequence 5'-TGAC-3'. {ECO:0000269|PubMed:17965274}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00675 | ChIP-seq | Transfer from GRMZM2G017087 | Download |
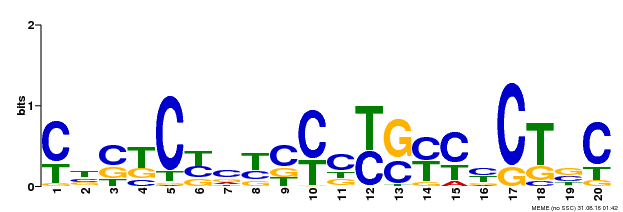
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | DQ317418 | 0.0 | DQ317418.1 Panicum miliaceum KNOTTED1 homeodomain protein (KN1) mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025795239.1 | 0.0 | homeotic protein knotted-1 | ||||
| Swissprot | P24345 | 0.0 | KN1_MAIZE; Homeotic protein knotted-1 | ||||
| TrEMBL | A0A2S3IIG3 | 0.0 | A0A2S3IIG3_9POAL; Uncharacterized protein | ||||
| TrEMBL | A0A2T7C1P5 | 0.0 | A0A2T7C1P5_9POAL; Uncharacterized protein | ||||
| STRING | GRMZM2G017087_P02 | 0.0 | (Zea mays) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP9886 | 31 | 45 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G08150.1 | 2e-89 | KNOTTED-like from Arabidopsis thaliana | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pahal.I01913.1 |



