 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pahal.I04672.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1026aa MW: 113946 Da PI: 5.4408 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 171.5 | 1.2e-53 | 21 | 138 | 2 | 118 |
CG-1 2 lke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfq 95
lke ++rwl++ ei++iL+n+++++++ e+++rp+sgsl+L++rk++ryfrkDG++w+kk d ktv+E+he+LK g+++vl+cyYah+e+n +fq
Pahal.I04672.1 21 LKEaQHRWLRPAEICEILKNYRNFRIAPEPPNRPPSGSLFLFDRKVLRYFRKDGHNWRKKNDQKTVKEAHERLKSGSIDVLHCYYAHGEDNINFQ 115
45559****************************************************************************************** PP
CG-1 96 rrcywlLeeelekivlvhylevk 118
rr+yw+Lee++ +ivlvhyle+k
Pahal.I04672.1 116 RRTYWMLEEDFMHIVLVHYLETK 138
********************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 80.649 | 17 | 143 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 1.8E-72 | 20 | 138 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 5.2E-48 | 23 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 4.9E-6 | 446 | 533 | IPR013783 | Immunoglobulin-like fold |
| SuperFamily | SSF81296 | 1.58E-16 | 447 | 533 | IPR014756 | Immunoglobulin E-set |
| Pfam | PF01833 | 2.1E-6 | 447 | 525 | IPR002909 | IPT domain |
| SuperFamily | SSF48403 | 7.93E-16 | 618 | 741 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 1.6E-16 | 642 | 743 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 1.47E-11 | 642 | 739 | No hit | No description |
| PROSITE profile | PS50297 | 15.937 | 648 | 713 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 8.683 | 648 | 680 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 8.736 | 681 | 713 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 1.78E-8 | 840 | 893 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 0.086 | 842 | 864 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.407 | 843 | 872 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0032 | 844 | 863 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0022 | 865 | 887 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.56 | 866 | 890 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 2.0E-4 | 868 | 887 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1026 aa Download sequence Send to blast |
MASAEARRLA VVPKLDIEQI LKEAQHRWLR PAEICEILKN YRNFRIAPEP PNRPPSGSLF 60 LFDRKVLRYF RKDGHNWRKK NDQKTVKEAH ERLKSGSIDV LHCYYAHGED NINFQRRTYW 120 MLEEDFMHIV LVHYLETKGG KSSRARVNNN MIQEAAVDSP LSQLPSQTIE GESSLSGQAS 180 EYEEAESDIY SGGAGYHSFT RVQQHENGTG PVIDSSVFSS YTPASSIGNY QGLHAMTQNT 240 SFYPGNQHSS LVLNGSSTGV ATDGYANQTD LTSWNPVIEL DNGPVQMPLQ FPVPPEQGTS 300 TEGLGIDYLT FDEVYSDGLS LKDIGAAGAG GESFWQFPSA TGDLSTAENS FPQPNDDSLE 360 AAIGYPFLKT QSSNLSDILK DSFKKTDSFT RWMSKELLEV EDSQIQSSSG AYWSTEQADS 420 IIEASSREPL DQFTVSPMLS QDQLFSIVDF APTWTYVGSK TKILVTGSFL NNSQVTEKCK 480 WSCMFGEVEI PAKILADGTL LCYSPQHKPG RVPFYITCSN RLACSEVREF EFRPTVTQYM 540 DAPSPHGATN KVYFQIRLDK LLSLGPDEYQ ATVSNPSLEM IELSKKISSL MTTNDEWSNL 600 LKLACDNEPS TDDQQDQFAE NLIKNKLHVW LLNKVGVGGK GPSVLDDEGQ GVLHLAAALG 660 YDWAIRPTLA AGVNINFRDV HGWTALHWAA FCGRERTVVA LIALGAAPGA LTDPSPDFPE 720 STPADLASAN GQKGISGFLA ESSLTSHLQA LNLKEANMAE ISGLPGIGDV TERDSLQPPS 780 GDSLGPVRNA AQAAARIYQV FRVQSFQRKQ AAQYEDDKGG LSDERALSLL SVKPSKPGQL 840 DPLHSAATRI QNKFRGWKGR KEFLLIRQRI VKIQAHVRGH QVRKHYRKIV WSVGIVEKVI 900 LRWRRRGAGL RGFRSTEGSS EGSSGGTSSN MIKDKPSGDD YDFLQEGRKQ TEERLQKALA 960 RVKSMAEYPE ARDQYQRILT VVSKMQESQA MQEKMLEEST EMDDGYFMNE LQELWDDDTP 1020 LPGYF* |
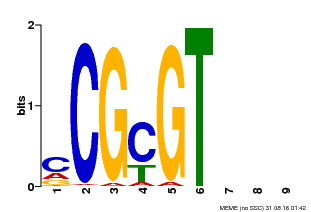
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00042 | PBM | Transfer from AT2G22300 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025793388.1 | 0.0 | calmodulin-binding transcription activator 3-like isoform X2 | ||||
| TrEMBL | A0A2S3IMM7 | 0.0 | A0A2S3IMM7_9POAL; Uncharacterized protein | ||||
| STRING | Pavir.Ia02749.1.p | 0.0 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP608 | 38 | 140 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G64220.2 | 0.0 | Calmodulin-binding transcription activator protein with CG-1 and Ankyrin domains | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pahal.I04672.1 |



