 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pahal.J01902.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 274aa MW: 29252.3 Da PI: 4.5207 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 60.2 | 3.4e-19 | 48 | 101 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k++++++eq+++Le+ Fe ++++ e++++LA+ lgL+ rqV vWFqNrRa++k
Pahal.J01902.1 48 KKRRLSAEQVRALERSFEVENKLEPERKARLARDLGLQPRQVAVWFQNRRARWK 101
45689************************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 134.3 | 4.2e-43 | 47 | 139 | 1 | 93 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreelke 93
ekkrrls+eqv++LE+sFe e+kLeperK++lar+LglqprqvavWFqnrRAR+ktkqlE+dy+aL+++ydal++++++L++++++L +e+ke
Pahal.J01902.1 47 EKKRRLSAEQVRALERSFEVENKLEPERKARLARDLGLQPRQVAVWFQNRRARWKTKQLERDYAALRHSYDALRHDHDALRRDKDALLAEIKE 139
69**************************************************************************************99987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.21E-19 | 41 | 105 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.475 | 43 | 103 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.5E-18 | 46 | 107 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 1.6E-16 | 48 | 101 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 6.92E-18 | 48 | 104 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 2.9E-20 | 49 | 110 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 9.3E-6 | 74 | 83 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 78 | 101 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 9.3E-6 | 83 | 99 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 1.3E-17 | 103 | 144 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009637 | Biological Process | response to blue light | ||||
| GO:0030308 | Biological Process | negative regulation of cell growth | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0048510 | Biological Process | regulation of timing of transition from vegetative to reproductive phase | ||||
| GO:0048573 | Biological Process | photoperiodism, flowering | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 274 aa Download sequence Send to blast |
MKRPRGGSPS LLPTANPDDD DGYDAAAGME AEGDAEEMVA RGGGGGEKKR RLSAEQVRAL 60 ERSFEVENKL EPERKARLAR DLGLQPRQVA VWFQNRRARW KTKQLERDYA ALRHSYDALR 120 HDHDALRRDK DALLAEIKEL KAKLGDEEAA ASFTSVKAEP AASDGPPVAG VGSSESDSSA 180 VLNDADAPAG VAEAPVPEVQ GALLPTPAAA AGAATSHGGA FFHGGFLKVE EDETGFLDDD 240 EPCGGFFAVE QPPPMAWWTE PTEHCLLELN WGG* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 72 | 78 | ERKARLA |
| 2 | 95 | 103 | RRARWKTKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription activator that binds to the DNA sequence 5'-CAAT[AT]ATTG-3'. May be involved in the regulation of gibberellin signaling. {ECO:0000269|PubMed:10732669, ECO:0000269|PubMed:18049796}. | |||||
| UniProt | Probable transcription activator that binds to the DNA sequence 5'-CAAT[AT]ATTG-3'. May be involved in the regulation of gibberellin signaling. {ECO:0000269|PubMed:10732669, ECO:0000269|PubMed:18049796}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00051 | PBM | Transfer from AT4G40060 | Download |
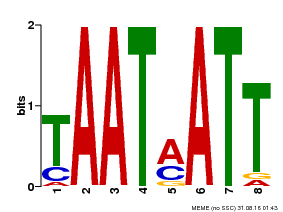
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By gibberellin. Down-regulated in leaves by drought stress. {ECO:0000269|PubMed:17999151, ECO:0000269|PubMed:18049796}. | |||||
| UniProt | INDUCTION: By gibberellin. Down-regulated in leaves by drought stress. {ECO:0000269|PubMed:17999151, ECO:0000269|PubMed:18049796}. | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT034257 | 0.0 | BT034257.1 Zea mays full-length cDNA clone ZM_BFc0009H14 mRNA, complete cds. | |||
| GenBank | BT039062 | 0.0 | BT039062.1 Zea mays full-length cDNA clone ZM_BFb0367N11 mRNA, complete cds. | |||
| GenBank | BT055374 | 0.0 | BT055374.1 Zea mays full-length cDNA clone ZM_BFb0091J03 mRNA, complete cds. | |||
| GenBank | JX469929 | 0.0 | JX469929.1 Zea mays subsp. mays clone UT3187 HB-type transcription factor mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025803235.1 | 0.0 | homeobox-leucine zipper protein HOX4-like | ||||
| Swissprot | Q6K498 | 1e-121 | HOX4_ORYSJ; Homeobox-leucine zipper protein HOX4 | ||||
| Swissprot | Q9XH37 | 1e-121 | HOX4_ORYSI; Homeobox-leucine zipper protein HOX4 | ||||
| TrEMBL | A0A2S3H0J4 | 0.0 | A0A2S3H0J4_9POAL; Uncharacterized protein | ||||
| STRING | Si030822m | 1e-162 | (Setaria italica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3009 | 34 | 82 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G40060.1 | 2e-40 | homeobox protein 16 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pahal.J01902.1 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



