 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pavir.1KG020700.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 870aa MW: 98182.9 Da PI: 8.067 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 13 | 0.0003 | 557 | 579 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
++C Cg +F + +Lk+H+++H
Pavir.1KG020700.1.p 557 HRCLECGACFQKPAHLKQHMQSH 579
79*******************99 PP
| |||||||
| 2 | zf-C2H2 | 20.4 | 1.3e-06 | 585 | 609 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
+ C+ dC+ s++rk++L+rH+ +H
Pavir.1KG020700.1.p 585 FICQleDCPFSYKRKDHLNRHMLKH 609
789999*****************99 PP
| |||||||
| 3 | zf-C2H2 | 13.1 | 0.00029 | 614 | 639 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
++C+ C+++Fs k n++rH + H
Pavir.1KG020700.1.p 614 FSCTvdGCDRRFSIKANMQRHVKEfH 639
899999****************9988 PP
| |||||||
| 4 | zf-C2H2 | 15.2 | 6.1e-05 | 658 | 682 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
+ C+ C+k F+ s Lk+H +H
Pavir.1KG020700.1.p 658 FICKeeGCNKAFKYLSKLKKHEESH 682
789999***************9988 PP
| |||||||
| 5 | zf-C2H2 | 18.2 | 6.7e-06 | 749 | 773 | 2 | 23 |
EET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 2 kCp..dCgksFsrksnLkrHirt.H 23
kC+ C++sFs+ksnL++H + H
Pavir.1KG020700.1.p 749 KCTfdGCDRSFSSKSNLTKHKKAcH 773
799999***************9955 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.25.40.10 | 1.8E-7 | 28 | 163 | IPR011990 | Tetratricopeptide-like helical domain |
| PROSITE profile | PS51375 | 5.261 | 35 | 65 | IPR002885 | Pentatricopeptide repeat |
| PROSITE profile | PS51375 | 9.065 | 66 | 100 | IPR002885 | Pentatricopeptide repeat |
| Pfam | PF01535 | 0.46 | 68 | 97 | IPR002885 | Pentatricopeptide repeat |
| PROSITE profile | PS51375 | 8.046 | 136 | 166 | IPR002885 | Pentatricopeptide repeat |
| Pfam | PF01535 | 0.0022 | 141 | 163 | IPR002885 | Pentatricopeptide repeat |
| PROSITE profile | PS51375 | 9.832 | 167 | 201 | IPR002885 | Pentatricopeptide repeat |
| Pfam | PF01535 | 0.017 | 169 | 197 | IPR002885 | Pentatricopeptide repeat |
| TIGRFAMs | TIGR00756 | 1.4E-4 | 200 | 233 | IPR002885 | Pentatricopeptide repeat |
| Pfam | PF01535 | 4.2E-4 | 200 | 229 | IPR002885 | Pentatricopeptide repeat |
| PROSITE profile | PS51375 | 6.434 | 202 | 232 | IPR002885 | Pentatricopeptide repeat |
| Gene3D | G3DSA:1.25.40.10 | 1.8E-7 | 261 | 335 | IPR011990 | Tetratricopeptide-like helical domain |
| PROSITE profile | PS51375 | 8.605 | 268 | 298 | IPR002885 | Pentatricopeptide repeat |
| TIGRFAMs | TIGR00756 | 0.0011 | 273 | 298 | IPR002885 | Pentatricopeptide repeat |
| Pfam | PF13041 | 6.6E-11 | 298 | 346 | IPR002885 | Pentatricopeptide repeat |
| PROSITE profile | PS51375 | 12.09 | 299 | 333 | IPR002885 | Pentatricopeptide repeat |
| TIGRFAMs | TIGR00756 | 3.5E-7 | 301 | 335 | IPR002885 | Pentatricopeptide repeat |
| PROSITE profile | PS51375 | 9.021 | 334 | 364 | IPR002885 | Pentatricopeptide repeat |
| TIGRFAMs | TIGR00756 | 0.001 | 336 | 369 | IPR002885 | Pentatricopeptide repeat |
| PROSITE profile | PS51375 | 6.774 | 370 | 400 | IPR002885 | Pentatricopeptide repeat |
| Pfam | PF01535 | 0.05 | 373 | 397 | IPR002885 | Pentatricopeptide repeat |
| PROSITE profile | PS51375 | 5.196 | 402 | 432 | IPR002885 | Pentatricopeptide repeat |
| Gene3D | G3DSA:1.25.40.10 | 1.8E-7 | 403 | 464 | IPR011990 | Tetratricopeptide-like helical domain |
| PROSITE profile | PS51375 | 7.158 | 436 | 470 | IPR002885 | Pentatricopeptide repeat |
| SMART | SM00355 | 31 | 511 | 533 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 6.2E-4 | 554 | 576 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 12.591 | 557 | 584 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.04 | 557 | 579 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 559 | 579 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 1.49E-10 | 569 | 611 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 6.6E-11 | 577 | 611 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 9.8E-4 | 585 | 609 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 9.078 | 585 | 614 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 587 | 609 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 2.15E-7 | 595 | 646 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 5.9E-8 | 612 | 640 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0036 | 614 | 639 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.762 | 614 | 644 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 616 | 639 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 9.7E-7 | 654 | 682 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0087 | 658 | 682 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.201 | 658 | 682 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 660 | 682 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 8.2 | 690 | 715 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 692 | 715 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 13 | 718 | 739 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 1.9E-6 | 733 | 770 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 5.99E-8 | 733 | 790 | No hit | No description |
| PROSITE profile | PS50157 | 11.177 | 748 | 778 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 6.0E-4 | 748 | 773 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 750 | 773 | IPR007087 | Zinc finger, C2H2 |
| PROSITE profile | PS50157 | 8.933 | 779 | 805 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 1.3 | 779 | 805 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 781 | 805 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0008097 | Molecular Function | 5S rRNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0080084 | Molecular Function | 5S rDNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 870 aa Download sequence Send to blast |
MNRSICHHLL SLCKTLRELQ KLHAQAVAQG LHPRHQSVSC KIFRCYADFG RAADARKLFD 60 EIPRPDLVSF TSLMSLHIRL ERHRAALSLF SRAVADGHRP DGFAVVGALS ASGGAGDHQV 120 GRAVHGLIFR LGLHSEVVVG NALVDMYSRC GKFESALLVF DRMFLKDEIT WGSMLHGYIK 180 CAGFDLALSF FDQMPVKSVV AWTALITGHV QVRLPVRALE LFGRMVLEGH CPTHVTIVGV 240 LSACADIGAL DLGRIIHGYG IKCNASSNII VSNALMDMYA KSGSVEMAFS VFQEVQSKDA 300 FTWTTMISCF TVQGDGRKAL ELFWDMLRSG VVPNSVTFVS VLSACSHAGL IEEGRQLFGT 360 MREMYGIDPQ LEHYGCMVDL LARGGMLEEA EALIVDMNVE PDIVIWRSLL SACLVHRNAR 420 LAEIAGKEIM KREPGDDGVY VLLWNMYASS NKWREARAIR QQMLTRKIFK KPGCSWIEVD 480 GVVHEFLMCS GDEIDGDASV GHNGFKDIRR YNCEFCTVVR SKKCLIQAHM VEHHKDELDK 540 SETYNSNGEK IVCEEEHRCL ECGACFQKPA HLKQHMQSHS NERLFICQLE DCPFSYKRKD 600 HLNRHMLKHQ GKLFSCTVDG CDRRFSIKAN MQRHVKEFHD DVKEFHDNEN VTKSNQQFIC 660 KEEGCNKAFK YLSKLKKHEE SHVKLNYTEV VCCEPGCMKM FTNVECLRAH NQSCHQHVQC 720 EICGKKHLKK NIKRHLQAHD EVASGERIKC TFDGCDRSFS SKSNLTKHKK ACHDRMKHFT 780 CRVAGCGKVF TYKHVRDNHE KSSAHVYVEG DFEEMDEQLR SRPRGGRKRK ALTVETLTRK 840 RVTIPGEASS LLDGAEYLSW LLSGGDEAQ* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5iww_D | 2e-59 | 20 | 417 | 3 | 338 | PLS9-PPR |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 821 | 829 | RPRGGRKRK |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Pvr.19738 | 0.0 | callus| leaf| root| stem | ||||
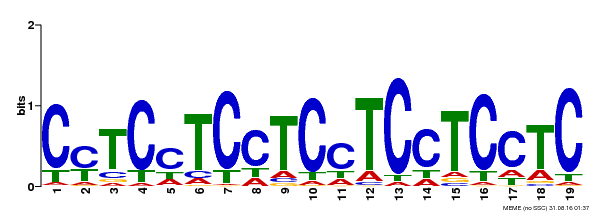
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00229 | DAP | Transfer from AT1G72050 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pavir.1KG020700.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025795590.1 | 0.0 | pentatricopeptide repeat-containing protein At2g22410, mitochondrial-like | ||||
| TrEMBL | K3Z0X6 | 0.0 | K3Z0X6_SETIT; Uncharacterized protein | ||||
| STRING | Pavir.Ab00169.1.p | 0.0 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2382 | 38 | 82 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G72050.2 | 1e-116 | transcription factor IIIA | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pavir.1KG020700.1.p |



