 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pavir.1NG436700.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 345aa MW: 36417.3 Da PI: 4.6902 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 53.3 | 6.4e-17 | 16 | 63 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg+WT++Ed +lv +++++G +W++ ++ g+ R++k+c++rw +yl
Pavir.1NG436700.1.p 16 RGSWTPQEDMRLVAYIQKHGHSNWRALPKQAGLLRCGKSCRLRWINYL 63
89******************************99************97 PP
| |||||||
| 2 | Myb_DNA-binding | 51.1 | 3e-16 | 69 | 114 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg++T eE+e +++++ +lG++ W++Ia++++ gRt++++k+ w+++l
Pavir.1NG436700.1.p 69 RGNFTVEEEETIIRLHGMLGNK-WSKIAACLP-GRTDNEIKNVWNTHL 114
89********************.*********.************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 14.227 | 11 | 63 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.85E-30 | 13 | 110 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 8.3E-13 | 15 | 65 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.0E-15 | 16 | 63 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.2E-23 | 17 | 70 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 2.68E-10 | 18 | 63 | No hit | No description |
| PROSITE profile | PS51294 | 24.896 | 64 | 118 | IPR017930 | Myb domain |
| SMART | SM00717 | 2.1E-14 | 68 | 116 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.1E-14 | 69 | 114 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 9.89E-11 | 71 | 114 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 4.1E-26 | 71 | 118 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:2000652 | Biological Process | regulation of secondary cell wall biogenesis | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 345 aa Download sequence Send to blast |
MGRGRAPCCA KVGLNRGSWT PQEDMRLVAY IQKHGHSNWR ALPKQAGLLR CGKSCRLRWI 60 NYLRPDLKRG NFTVEEEETI IRLHGMLGNK WSKIAACLPG RTDNEIKNVW NTHLKKKVAQ 120 REQQKKAGAA KNDGAASGGD AGTPGTDSSS ASSSTTTTTT NNSSGGSDSG DQCGTSKEPD 180 AAAVSPLHDI GVWEMLVDAP AAAQPMLSSS CSSSSLTTCV GGAEDLIELP VMDIEPDIWS 240 IIDDESADGS GARHGDAAAP CTGAAVSTSE EEAGEAADDW WLENLEKELG LWGPTEDPLA 300 HSDLLVDHTG FGPLSSSEGG PVSTYFQSGP DNAATEQLLG RVWL* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h8a_C | 1e-26 | 14 | 118 | 25 | 128 | MYB TRANSFORMING PROTEIN |
| 1mse_C | 1e-26 | 14 | 118 | 2 | 105 | C-Myb DNA-Binding Domain |
| 1msf_C | 1e-26 | 14 | 118 | 2 | 105 | C-Myb DNA-Binding Domain |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that regulates positively genes involved in anthocyanin biosynthesis such as A1. {ECO:0000269|PubMed:7920701}. | |||||
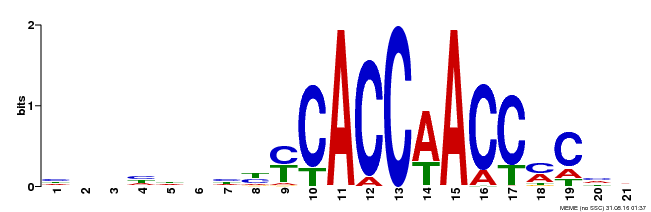
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00249 | DAP | Transfer from AT1G79180 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pavir.1NG436700.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HF679437 | 0.0 | HF679437.1 Saccharum hybrid cultivar Co 86032 mRNA for ScMYB31 protein. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025814304.1 | 0.0 | myb-related protein Zm1-like | ||||
| Swissprot | P20024 | 1e-159 | MYB1_MAIZE; Myb-related protein Zm1 | ||||
| TrEMBL | A0A2S3GS79 | 1e-180 | A0A2S3GS79_9POAL; Uncharacterized protein | ||||
| STRING | Pavir.Aa01159.1.p | 0.0 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2154 | 37 | 98 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G79180.1 | 3e-68 | myb domain protein 63 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pavir.1NG436700.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



