 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pavir.2KG384800.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 246aa MW: 27373.5 Da PI: 8.811 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 54.6 | 2.5e-17 | 14 | 61 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEd++lv ++k +G g+W++ ++ g+ R++k+c++rw +yl
Pavir.2KG384800.1.p 14 KGAWTKEEDDRLVAYIKAHGEGCWRSLPKAAGLLRCGKSCRLRWINYL 61
79******************************99************97 PP
| |||||||
| 2 | Myb_DNA-binding | 60.2 | 4.3e-19 | 67 | 111 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
rg++T+eEdel+++++ +lG++ W++Ia +++ gRt++++k++w+++
Pavir.2KG384800.1.p 67 RGNFTEEEDELIIKLHSLLGNK-WSLIAGRLP-GRTDNEIKNYWNTH 111
89********************.*********.************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 6.5E-24 | 5 | 64 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 16.58 | 9 | 61 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.9E-13 | 13 | 63 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 5.64E-30 | 13 | 108 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 4.3E-15 | 14 | 61 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 4.93E-10 | 16 | 61 | No hit | No description |
| PROSITE profile | PS51294 | 29.411 | 62 | 116 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.6E-28 | 65 | 116 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 6.3E-18 | 66 | 114 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.9E-17 | 67 | 111 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 7.29E-13 | 69 | 112 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0010224 | Biological Process | response to UV-B | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:1903086 | Biological Process | negative regulation of sinapate ester biosynthetic process | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 246 aa Download sequence Send to blast |
MGRSPCCEKA HTNKGAWTKE EDDRLVAYIK AHGEGCWRSL PKAAGLLRCG KSCRLRWINY 60 LRPDLKRGNF TEEEDELIIK LHSLLGNKWS LIAGRLPGRT DNEIKNYWNT HIRRKLLSRG 120 IDPVSHRPIN EHASNITISF EAAAAAREEK GAVFRLEEPK AAAAIGRDQN PGGDWGHGKP 180 LKCPDLNLDL CISPPCQEEP LKPVKREAGL CFSCSLGLPK SAECKCSSFL GLRTAMLDFR 240 SLEMK* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h8a_C | 1e-29 | 13 | 116 | 26 | 128 | MYB TRANSFORMING PROTEIN |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Pvr.25863 | 1e-130 | root | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Germinating seed and apical meristem of shoot and root. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Possible transcription activator in response to an external signal. May be involved in the regulation of flavonoid biosynthesis. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00479 | DAP | Transfer from AT4G38620 | Download |
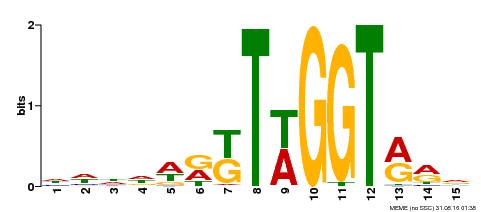
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pavir.2KG384800.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HF679410 | 0.0 | HF679410.1 Saccharum hybrid cultivar Co 86032 mRNA for ScMYB4 protein. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025803176.1 | 1e-168 | myb-related protein Hv1-like | ||||
| Swissprot | P20026 | 1e-138 | MYB1_HORVU; Myb-related protein Hv1 | ||||
| TrEMBL | A0A2S3H1Q9 | 1e-167 | A0A2S3H1Q9_9POAL; Uncharacterized protein | ||||
| STRING | Pavir.Bb02469.1.p | 1e-180 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP116 | 37 | 448 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G38620.1 | 1e-101 | myb domain protein 4 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pavir.2KG384800.1.p |



