 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pavir.2KG507900.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 318aa MW: 33741.9 Da PI: 8.813 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 205.7 | 1.3e-63 | 12 | 153 | 2 | 141 |
DUF822 2 gsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeaeaagssasaspe 91
g gr+ptwkErEnnkrRERrRRaiaaki++GLRa Gnyklpk++DnneVlkALcreAGwvvedDGttyrkg+kp+ +sp+
Pavir.2KG507900.1.p 12 GLGRTPTWKERENNKRRERRRRAIAAKIFTGLRALGNYKLPKHCDNNEVLKALCREAGWVVEDDGTTYRKGCKPP--------PGMMSPC 93
679************************************************************************........4557889 PP
DUF822 92 sslq.sslkssalaspvesysaspksssfpspssldsislasa.........asllpvls 141
ss+q +s++ss+ +spv+sy+asp+sssfpsp++ld s +++ +sllp+l+
Pavir.2KG507900.1.p 94 SSSQlLSATSSSYPSPVPSYHASPASSSFPSPTRLDPSSGSNThnpgaaaavSSLLPFLR 153
998888999999*************************99987766666666666666655 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 4.1E-61 | 13 | 154 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009742 | Biological Process | brassinosteroid mediated signaling pathway | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0048316 | Biological Process | seed development | ||||
| GO:0048481 | Biological Process | plant ovule development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005829 | Cellular Component | cytosol | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 318 aa Download sequence Send to blast |
MTSGAAVAAT GGLGRTPTWK ERENNKRRER RRRAIAAKIF TGLRALGNYK LPKHCDNNEV 60 LKALCREAGW VVEDDGTTYR KGCKPPPGMM SPCSSSQLLS ATSSSYPSPV PSYHASPASS 120 SFPSPTRLDP SSGSNTHNPG AAAAVSSLLP FLRGLPNLPP LRVSSSAPVT PPLSSPTAAS 180 RPPTKVRKPD WDAVAVDPFR HPFFAVSAPA SPTRARRREH PDTIPECDES DVCSTVDSGR 240 WISFQMSAAT TAPASPTYNL VNPSSGASAS TSMELDGMAA AEFEFDKGRV TPWEGERIHE 300 VAAEELELTL TLSVGAK* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5zd4_A | 5e-28 | 15 | 95 | 372 | 452 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_B | 5e-28 | 15 | 95 | 372 | 452 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_C | 5e-28 | 15 | 95 | 372 | 452 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_D | 5e-28 | 15 | 95 | 372 | 452 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Pvr.14492 | 0.0 | root| stem | ||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Positive brassinosteroid-signaling protein. Mediates downstream brassinosteroid-regulated growth response and feedback inhibition of brassinosteroid biosynthetic genes. May act as transcriptional repressor by binding the brassinosteroid-response element (5'-CGTGCG-3') in the promoter of GRAS32 (AC Q9LWU9), another positive regulator of brassinosteroid signaling (By similarity). {ECO:0000250}. | |||||
| UniProt | Positive brassinosteroid-signaling protein. Mediates downstream brassinosteroid-regulated growth response and feedback inhibition of brassinosteroid (BR) biosynthetic genes (PubMed:17699623, PubMed:19220793). May act as transcriptional repressor by binding the brassinosteroid-response element (BREE) (5'-CGTG(T/C)G-3') in the promoter of DLT (AC Q9LWU9), another positive regulator of BR signaling (PubMed:19220793). Acts as transcriptional repressor of LIC, a negative regulator of BR signaling, by binding to the BRRE element of its promoter. BZR1 and LIC play opposite roles in BR signaling and regulation of leaf bending (PubMed:22570626). {ECO:0000269|PubMed:17699623, ECO:0000269|PubMed:19220793, ECO:0000269|PubMed:22570626}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00237 | DAP | Transfer from AT1G75080 | Download |
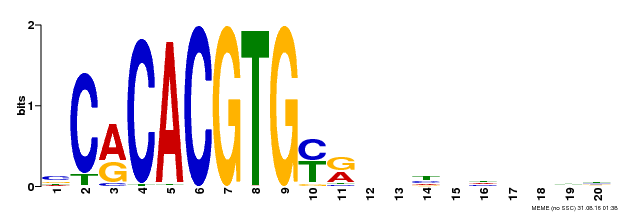
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pavir.2KG507900.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated by 24-epibrassinolide. {ECO:0000269|PubMed:22570626}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EU969826 | 0.0 | EU969826.1 Zea mays clone 336150 BES1/BZR1 protein mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025802391.1 | 1e-168 | protein BZR1 homolog 1-like | ||||
| Swissprot | B8B7S5 | 1e-135 | BZR1_ORYSI; Protein BZR1 homolog 1 | ||||
| Swissprot | Q7XI96 | 1e-135 | BZR1_ORYSJ; Protein BZR1 homolog 1 | ||||
| TrEMBL | A0A3L6TE71 | 1e-169 | A0A3L6TE71_PANMI; Uncharacterized protein | ||||
| STRING | Pavir.Bb03208.1.p | 0.0 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP5474 | 34 | 53 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G19350.3 | 2e-43 | BES1 family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pavir.2KG507900.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



