 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pavir.2KG511300.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 578aa MW: 61619.6 Da PI: 6.5105 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 101.3 | 5.6e-32 | 195 | 252 | 2 | 60 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS-- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhek 60
+DgynWrKYGqK+vkgse+prsYY+Ct+++C+vkk +ers d++++e++Y+g Hnh+k
Pavir.2KG511300.1.p 195 EDGYNWRKYGQKHVKGSENPRSYYKCTHPNCEVKKLLERSL-DGQITEVVYKGLHNHPK 252
8****************************************.***************85 PP
| |||||||
| 2 | WRKY | 107.5 | 6.4e-34 | 368 | 426 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+vkg+++prsYY+Ct++gCpv+k+ver+++dpk v++tYeg+Hnhe
Pavir.2KG511300.1.p 368 LDDGYRWRKYGQKVVKGNPNPRSYYKCTHTGCPVRKHVERASHDPKSVITTYEGKHNHE 426
59********************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 1.2E-27 | 185 | 253 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 23.296 | 189 | 253 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 7.98E-25 | 193 | 253 | IPR003657 | WRKY domain |
| SMART | SM00774 | 4.4E-35 | 194 | 252 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 3.7E-24 | 195 | 251 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 5.6E-38 | 353 | 428 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 5.36E-30 | 360 | 428 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 38.297 | 363 | 428 | IPR003657 | WRKY domain |
| SMART | SM00774 | 6.5E-40 | 368 | 427 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.2E-26 | 369 | 426 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009961 | Biological Process | response to 1-aminocyclopropane-1-carboxylic acid | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 578 aa Download sequence Send to blast |
MADSPNPSSG DLPAGSGGSP EKPVLADRRV AALAGAGAGA RYKAMSPARL PISREPCLTI 60 PAGFSPGALL ESPVLLNNFK VEPSPTTGTL SMAAIINKST HLDILPSPRE KSAGSGNEDG 120 DSRDFEFKPH LSCQPAESIH QTANISSVPN QPVAIVGPSD KMPAEVGTSE MNQMNSSENA 180 AQETQTENVA EKSAEDGYNW RKYGQKHVKG SENPRSYYKC THPNCEVKKL LERSLDGQIT 240 EVVYKGLHNH PKPQPNRRLA AGAVPSSQGE ERYDGAAPVE DKPSNIYPNL CNQVHSAGMI 300 DPVPGPASDD DGDGGGGRSY PGDDANEDDD LDSKRRKMES AGIDAALMGK PNREPRVVVQ 360 TVSEVDILDD GYRWRKYGQK VVKGNPNPRS YYKCTHTGCP VRKHVERASH DPKSVITTYE 420 GKHNHEVPAS RNASHEMSAA PMKPAVHPIN SSMPGLGGMM RACDARAFTN QYSQAAESDT 480 ISLDLGVGIS PNHSDATNHM QPSVPEPMQY QMQHMAPVYG SMGFPGMPVP TVPGNAASSV 540 YGSREEKANE GFTFKATPLG RSANLCYSSA GNLVMGP* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 3e-36 | 195 | 429 | 8 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 3e-36 | 195 | 429 | 8 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Pvr.6246 | 0.0 | callus| leaf| root| stem | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Peak of expression 12 days after pollination. {ECO:0000269|PubMed:12953112}. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in endosperm, but not in leaves. {ECO:0000269|PubMed:12953112}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in starch synthesis (PubMed:12953112). Acts as a transcriptional activator in sugar signaling (PubMed:16167901). Interacts specifically with the SURE and W-box elements, but not with the SP8a element (PubMed:12953112). {ECO:0000269|PubMed:12953112, ECO:0000269|PubMed:16167901}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00455 | DAP | Transfer from AT4G26640 | Download |
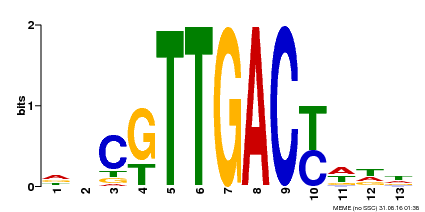
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pavir.2KG511300.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by sugar. {ECO:0000269|PubMed:12953112}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT054888 | 0.0 | BT054888.1 Zea mays full-length cDNA clone ZM_BFc0109P23 mRNA, complete cds. | |||
| GenBank | BT061714 | 0.0 | BT061714.1 Zea mays full-length cDNA clone ZM_BFb0176N11 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025801894.1 | 0.0 | WRKY transcription factor SUSIBA2-like isoform X1 | ||||
| Swissprot | Q6VWJ6 | 0.0 | WRK46_HORVU; WRKY transcription factor SUSIBA2 | ||||
| TrEMBL | A0A3L6QQW0 | 0.0 | A0A3L6QQW0_PANMI; WRKY transcription factor SUSIBA2-like | ||||
| STRING | Pavir.Bb03225.1.p | 0.0 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6740 | 38 | 51 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G26640.2 | 1e-118 | WRKY family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pavir.2KG511300.1.p |



