 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pavir.2NG407700.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 367aa MW: 39589.8 Da PI: 9.265 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 180 | 6e-56 | 23 | 153 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpkkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdk 90
lppGfrFhPtdeel+++yL++k+++ ++++ ++i+evd++k+ePwdLp+k+k +ekewyfFs rd+ky+tg r+nrat++gyWk+tgkdk
Pavir.2NG407700.1.p 23 LPPGFRFHPTDEELITYYLRQKIADGSFTA-RAIAEVDLNKCEPWDLPEKAKLGEKEWYFFSLRDRKYPTGVRTNRATNAGYWKTTGKDK 111
79*************************999.89***************99999************************************* PP
NAM 91 evlsk....kgelvglkktLvfykgrapkgektdWvmheyrl 128
e+++ + elvg+kktLvfykgrap+gekt+Wvmheyrl
Pavir.2NG407700.1.p 112 EIYTGqlpaTPELVGMKKTLVFYKGRAPRGEKTNWVMHEYRL 153
***9766666667***************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 1.11E-64 | 20 | 174 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 61.187 | 23 | 174 | IPR003441 | NAC domain |
| Pfam | PF02365 | 2.1E-29 | 24 | 153 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 367 aa Download sequence Send to blast |
MEGSAAAAGG GVAGGGSKKE ESLPPGFRFH PTDEELITYY LRQKIADGSF TARAIAEVDL 60 NKCEPWDLPE KAKLGEKEWY FFSLRDRKYP TGVRTNRATN AGYWKTTGKD KEIYTGQLPA 120 TPELVGMKKT LVFYKGRAPR GEKTNWVMHE YRLHSKSTPK SNKDEWVVCR VFAKSAGAKK 180 YPSNNAHSRS HHHPYTLDMV PPLLPTLLQH DPFARGPHYP YMTPADLAEL ARFARGTPGL 240 HPHIQPHPGT AAAYINPAAV APPFTLSGGL SLNIGASPAM PSPPLHGMSM AMSHQQIGPS 300 GAGTAGNHHQ VMASDHQQQM APAGLGGCVI GPGADAGFGA DAAGARYQSL DVEQLVERYW 360 PAGYQV* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 6e-57 | 22 | 180 | 16 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 6e-57 | 22 | 180 | 16 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 6e-57 | 22 | 180 | 16 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 6e-57 | 22 | 180 | 16 | 171 | NO APICAL MERISTEM PROTEIN |
| 3swm_A | 9e-57 | 22 | 180 | 19 | 174 | NAC domain-containing protein 19 |
| 3swm_B | 9e-57 | 22 | 180 | 19 | 174 | NAC domain-containing protein 19 |
| 3swm_C | 9e-57 | 22 | 180 | 19 | 174 | NAC domain-containing protein 19 |
| 3swm_D | 9e-57 | 22 | 180 | 19 | 174 | NAC domain-containing protein 19 |
| 3swp_A | 9e-57 | 22 | 180 | 19 | 174 | NAC domain-containing protein 19 |
| 3swp_B | 9e-57 | 22 | 180 | 19 | 174 | NAC domain-containing protein 19 |
| 3swp_C | 9e-57 | 22 | 180 | 19 | 174 | NAC domain-containing protein 19 |
| 3swp_D | 9e-57 | 22 | 180 | 19 | 174 | NAC domain-containing protein 19 |
| 4dul_A | 6e-57 | 22 | 180 | 16 | 171 | NAC domain-containing protein 19 |
| 4dul_B | 6e-57 | 22 | 180 | 16 | 171 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Pvr.11073 | 0.0 | root | ||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds to the promoter regions of genes involved in chlorophyll catabolic processes, such as NYC1, SGR1, SGR2 and PAO. {ECO:0000269|PubMed:27021284}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00277 | DAP | Transfer from AT2G24430 | Download |
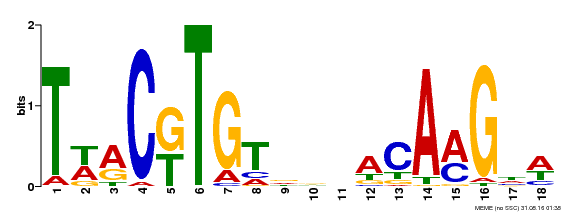
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pavir.2NG407700.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by the microRNA miR164. {ECO:0000269|PubMed:15294871, ECO:0000269|PubMed:17098808}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025803276.1 | 0.0 | NAC domain-containing protein 92-like | ||||
| Swissprot | Q9FLJ2 | 4e-84 | NC100_ARATH; NAC domain-containing protein 100 | ||||
| TrEMBL | A0A3L6QN81 | 0.0 | A0A3L6QN81_PANMI; Uncharacterized protein | ||||
| STRING | Pavir.Ba01665.1.p | 0.0 | (Panicum virgatum) | ||||
| STRING | Pavir.J02772.1.p | 0.0 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP9361 | 38 | 46 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G24430.2 | 2e-92 | NAC domain containing protein 38 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pavir.2NG407700.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



