 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pavir.2NG490000.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 915aa MW: 102606 Da PI: 8.5821 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 164.3 | 2.1e-51 | 33 | 147 | 4 | 117 |
CG-1 4 e.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenp 92
e +rw +++ei+a+L+n++++++++++ ++p sg+++Ly+rk+vr+frkDG++wkkkkdgktv+E+hekLK+g+ e +++yYa++ee+p
Pavir.2NG490000.1.p 33 EaGTRWFRPNEIYAVLANHARFKVHAQPIDKPASGTVVLYDRKVVRNFRKDGHNWKKKKDGKTVQEAHEKLKIGNEEKVHVYYARGEEDP 122
4379************************************************************************************** PP
CG-1 93 tfqrrcywlLeeelekivlvhylev 117
+f rrcywlL++ele+ivlvhy+++
Pavir.2NG490000.1.p 123 NFFRRCYWLLDKELERIVLVHYRQT 147
**********************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 72.702 | 27 | 153 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 6.0E-61 | 30 | 148 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 3.1E-44 | 34 | 146 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 9.38E-11 | 381 | 467 | IPR014756 | Immunoglobulin E-set |
| SuperFamily | SSF48403 | 5.29E-19 | 514 | 670 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 6.6E-19 | 516 | 672 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 9.92E-17 | 558 | 666 | No hit | No description |
| PROSITE profile | PS50297 | 18.404 | 574 | 678 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 4.1E-8 | 580 | 670 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 6.3E-6 | 607 | 636 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 11.808 | 607 | 639 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 520 | 646 | 675 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 2.42E-9 | 755 | 823 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 0.24 | 772 | 794 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.334 | 773 | 802 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0057 | 775 | 793 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0019 | 795 | 817 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 10.53 | 796 | 820 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 7.3E-4 | 797 | 817 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 7.3 | 876 | 898 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.249 | 878 | 906 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.12 | 879 | 898 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 915 aa Download sequence Send to blast |
MAGGAGARDP LVASEIHGFL TCADLNFDKL MAEAGTRWFR PNEIYAVLAN HARFKVHAQP 60 IDKPASGTVV LYDRKVVRNF RKDGHNWKKK KDGKTVQEAH EKLKIGNEEK VHVYYARGEE 120 DPNFFRRCYW LLDKELERIV LVHYRQTSEE NAIPPSHIEA EVAEVPPINV IHYTSPLTST 180 DSASARTELS SCAAAAAAPE EINSHGGRAI SCETDDHDSS LESFWADLLE SSMKNDTSVR 240 GGSLTPNQQT NYGMRDSGNN ILNTNATSNA ILYSPANVVS EAYAANPGLN QVSESYYGAL 300 KHQVNQSPSL STSDLDSQSK PHANSIMKTP VDSNMPSDVP ARQNSLGLWK SLDDDITCLV 360 NNPSSAIPIT RPVIDEKPFH IIEISSEWAY CTDDTKVLVV GYFLDNYKHL AGANLYCVIG 420 EQCVTADIVQ TGVYRFMARP HVPGRVNLYL TLDGKTPISE VLSFEYRIMP GSSDDDEPKR 480 SKLQMQMRLA RLLFSTSQKK IPPKFLVEGS RVSNLLSASA EKEWMDLFKY VIDSKGTNIP 540 ATESLLELVL RNKLQEWLVE KIIEGHKSTD RDDLGQGPIH LCSFLGYTWA IRLFSLSGFS 600 LDFRDSSGWT ALHWAAYYGR EKMVAALLSA GANPSLVTDP THDDPGGHTA ADLAARQGFD 660 GLAAYLAEKG LTAHFEAMSL SKDKRSTSRT QSIKQISKEY ENLTEQELCL RESLAAYRNA 720 ADAANNIQAA LRERTLKLQT KAIELANPEI EAATIVAAMR IQHAFRNYNR KKMMRAAARI 780 QSHFRTWQMR RNFMNMRRQA IKIQAAYRGH QVRRQYRKVI WSVGVVEKAI LRWRKKRKGL 840 RGIATGMPVA MSTDTEAAST AEEDYYQVGR QQAEDRFNRS VVRVQALFRS HRAQQEYRRM 900 KVAHEEAKVE FSQQ* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Pvr.9297 | 0.0 | callus| leaf| root| stem | ||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
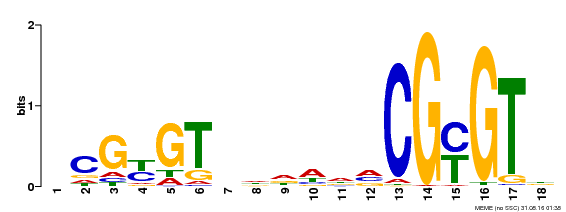
| UniProt | Transcription activator that binds calmodulin in a calcium-dependent manner in vitro. Binds to the DNA consensus sequence 5'-T[AC]CG[CT]GT[GT][GT][GT][GT]T[GT]CG-3'. {ECO:0000269|PubMed:16192280}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pavir.2NG490000.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025800750.1 | 0.0 | calmodulin-binding transcription activator CBT isoform X1 | ||||
| Swissprot | Q7XHR2 | 0.0 | CBT_ORYSJ; Calmodulin-binding transcription activator CBT | ||||
| TrEMBL | A0A3L6QU26 | 0.0 | A0A3L6QU26_PANMI; Calmodulin-binding transcription activator 6-like | ||||
| STRING | Pavir.Ba00948.1.p | 0.0 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP9831 | 29 | 33 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pavir.2NG490000.1.p |



