 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pavir.2NG588000.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 325aa MW: 36105.7 Da PI: 4.7756 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 58.4 | 1.7e-18 | 100 | 149 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
+y+G+r+++ +g+W+AeIrdp + r++lg+f taeeAa+a++ +++++g
Pavir.2NG588000.1.p 100 QYRGIRRRP-WGKWAAEIRDPQ----KgVRVWLGTFSTAEEAARAYDVEALRIRG 149
59*******.**********94....35************************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00380 | 6.7E-37 | 100 | 163 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 2.86E-31 | 100 | 159 | No hit | No description |
| SuperFamily | SSF54171 | 2.48E-22 | 100 | 158 | IPR016177 | DNA-binding domain |
| Gene3D | G3DSA:3.30.730.10 | 3.6E-32 | 100 | 158 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 24.961 | 100 | 157 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.3E-11 | 101 | 112 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 2.0E-11 | 101 | 149 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.3E-11 | 123 | 139 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009873 | Biological Process | ethylene-activated signaling pathway | ||||
| GO:0034059 | Biological Process | response to anoxia | ||||
| GO:0071456 | Biological Process | cellular response to hypoxia | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 325 aa Download sequence Send to blast |
MCGGAILSDL YSPVRRTVTA GDLWGESGSS KNVKNWKRSS WKFDEGDEDF EADFKDFEDC 60 SSEEEVDFGH EEKEFQLNSS NFVEFNGHTA KVTSRKRKIQ YRGIRRRPWG KWAAEIRDPQ 120 KGVRVWLGTF STAEEAARAY DVEALRIRGK KAKMNFPTTI TAAGKHHRQR VARPAKKTSQ 180 ESLKSSNASG HVISAGSSTD GTVVKIELSQ SPASPLPVSS AWLDAFELKQ LGGETPEADG 240 RETPEETDHE TGVTADMFFG NGEVRLSDDF ASYEPYPNFM QLPYLEGDSY ENIDTLFNGE 300 AAQDGVNIGG LWNFDDVPMD RGVY* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2gcc_A | 4e-21 | 100 | 162 | 5 | 68 | ATERF1 |
| 3gcc_A | 4e-21 | 100 | 162 | 5 | 68 | ATERF1 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Pvr.10332 | 0.0 | callus| root | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00230 | DAP | Transfer from AT1G72360 | Download |
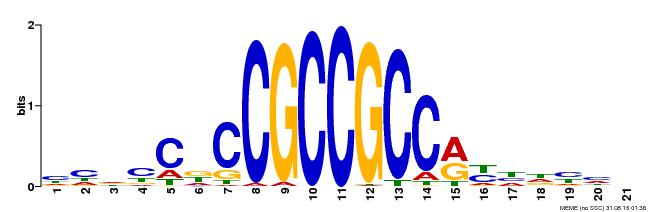
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pavir.2NG588000.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025801024.1 | 0.0 | ethylene-responsive transcription factor ERF073-like | ||||
| Refseq | XP_025801025.1 | 0.0 | ethylene-responsive transcription factor ERF073-like | ||||
| TrEMBL | A0A2S3H3X9 | 0.0 | A0A2S3H3X9_9POAL; Uncharacterized protein | ||||
| STRING | Pavir.Ba00410.1.p | 0.0 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP14560 | 21 | 23 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G72360.3 | 2e-37 | ERF family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pavir.2NG588000.1.p |



