 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pavir.2NG649800.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | HSF | ||||||||
| Protein Properties | Length: 305aa MW: 33826.9 Da PI: 6.9683 | ||||||||
| Description | HSF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HSF_DNA-bind | 112.6 | 2.7e-35 | 28 | 120 | 2 | 103 |
HHHHHHHHHCTGGGTTTSEESSSSSEEEES-HHHHHHHTHHHHSTT--HHHHHHHHHHTTEEE---SSBTTTTXTTSEEEEESXXXXXXX CS
HSF_DNA-bind 2 FlkklyeiledeelkeliswsengnsfvvldeeefakkvLpkyFkhsnfaSFvRQLnmYgFkkvkdeekkskskekiweFkhksFkkgkk 91
Fl+k+y++++d+ +++++sw e+ ++fvv+ + efa+++Lp+yFkh+nf+SFvRQLn+YgF+k+ ++ weF+++ F+kg k
Pavir.2NG649800.1.p 28 FLTKTYQLVDDPCTDHIVSWGEDDTTFVVWRPPEFARDLLPNYFKHNNFSSFVRQLNTYGFRKIVADR---------WEFANEFFRKGAK 108
9****************************************************************999.........************* PP
XXXXXXXXXXXX CS
HSF_DNA-bind 92 ellekikrkkse 103
+ll +i+r+ks+
Pavir.2NG649800.1.p 109 HLLAEIHRRKSA 120
*********986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.10 | 2.9E-37 | 20 | 111 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SMART | SM00415 | 5.1E-58 | 24 | 117 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| SuperFamily | SSF46785 | 1.8E-33 | 25 | 117 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| PRINTS | PR00056 | 1.1E-19 | 28 | 51 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| Pfam | PF00447 | 3.5E-31 | 28 | 117 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 1.1E-19 | 66 | 78 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PROSITE pattern | PS00434 | 0 | 67 | 91 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 1.1E-19 | 79 | 91 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0008356 | Biological Process | asymmetric cell division | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 305 aa Download sequence Send to blast |
MAFLVERCGE MVVSMESSPH AKPVPAPFLT KTYQLVDDPC TDHIVSWGED DTTFVVWRPP 60 EFARDLLPNY FKHNNFSSFV RQLNTYGFRK IVADRWEFAN EFFRKGAKHL LAEIHRRKSA 120 QPLPTPLPPH QPYHHLHHHH LNPFSPPPPP PTQPVYHFQE EPATAHGVHG GNNSDGSGGG 180 GDFLAALSED NRQLRRRNSL LLSELAHMKK LYNDIIYFLQ NHVAPVTSPS SAAHASQLPS 240 AGAASSCRLM ELDPGSPSPP PRPEAADADD GTVKLFGVAL QGKKKKRAHR EDGDDDHEQG 300 SSEV* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5d5u_B | 2e-21 | 24 | 117 | 26 | 129 | Heat shock factor protein 1 |
| 5d5v_B | 2e-21 | 24 | 117 | 26 | 129 | Heat shock factor protein 1 |
| 5d5v_D | 2e-21 | 24 | 117 | 26 | 129 | Heat shock factor protein 1 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Pvr.19854 | 1e-171 | root| stem | ||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that specifically binds DNA of heat shock promoter elements (HSE). {ECO:0000269|PubMed:16202242}. | |||||
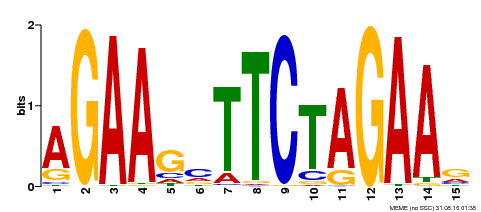
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00187 | DAP | Transfer from AT1G46264 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pavir.2NG649800.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT054148 | 0.0 | BT054148.1 Zea mays full-length cDNA clone ZM_BFb0133K13 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025804091.1 | 1e-175 | heat stress transcription factor B-4b-like | ||||
| Swissprot | Q7XHZ0 | 1e-131 | HFB4B_ORYSJ; Heat stress transcription factor B-4b | ||||
| TrEMBL | A0A1E5VCU8 | 0.0 | A0A1E5VCU8_9POAL; Heat stress transcription factor B-4b | ||||
| STRING | Pavir.J32257.1.p | 1e-172 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP121 | 37 | 394 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G46264.1 | 1e-63 | heat shock transcription factor B4 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pavir.2NG649800.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



