 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pavir.3KG079100.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 273aa MW: 31150.3 Da PI: 7.042 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 54.2 | 2.9e-17 | 131 | 194 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-ST...T---EEEEEE-SSSTT...EEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsa...gCpvkkkversaedpk...vveitYegeHnhe 59
Dg++WrKYGqK++k+s++ r YYrC+++ +C+++k v++++++++ + +++Y ++H+++
Pavir.3KG079100.1.p 131 YDGHQWRKYGQKTIKNSKHQRCYYRCAYKheqNCMATKTVQQQEHNTTesvMYTVVYYSHHTCK 194
59*************************99899********998876543338899******997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF118290 | 1.7E-19 | 124 | 194 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 2.6E-20 | 125 | 194 | IPR003657 | WRKY domain |
| SMART | SM00774 | 4.3E-28 | 130 | 195 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 2.5E-21 | 132 | 194 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 15.411 | 132 | 193 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009738 | Biological Process | abscisic acid-activated signaling pathway | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 273 aa Download sequence Send to blast |
MKLHTRYLPQ SSSTCADEFE HFACDHQPAI NEIVKVQSLV AQLQAIVLPA CEMMADLRSE 60 LVSQLFGNLQ DCTSKAISEL QTHHRAQMID IDRRRLAANI SDCKKVCAKK KSRINSRYGD 120 STSYMTPVPH YDGHQWRKYG QKTIKNSKHQ RCYYRCAYKH EQNCMATKTV QQQEHNTTES 180 VMYTVVYYSH HTCKANTDPA PPHIIETSTP QSAMMSSDSI IGSQEIVSPH TGSHKILENE 240 LATLQWTEDM QELLEKLADV PLDSDIWEMV RF* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6ir8_A | 9e-12 | 132 | 194 | 10 | 69 | OsWRKY45 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00066 | PBM | Transfer from AT1G66600 | Download |
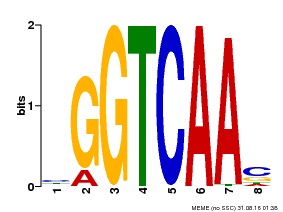
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pavir.3KG079100.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025805693.1 | 0.0 | probable WRKY transcription factor 63 | ||||
| TrEMBL | A0A2T7E6J3 | 1e-179 | A0A2T7E6J3_9POAL; Uncharacterized protein | ||||
| STRING | Pavir.Ca00459.1.p | 0.0 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3258 | 30 | 79 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G66600.1 | 3e-21 | ABA overly sensitive mutant 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pavir.3KG079100.1.p |



