 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pavir.3NG151500.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 417aa MW: 44331.7 Da PI: 6.9913 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 86 | 3.5e-27 | 224 | 283 | 2 | 58 |
--SS-EEEEEEE--TT-SS-EEEEEE-ST...T---EEEEEE-SSSTTEEEEEEES--SS CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsa...gCpvkkkversaedpkvveitYegeHnh 58
dDgy+WrKYGqK++ gs++prsYYrCt++ gC++kkkv+r ++dp + e+tY g+H++
Pavir.3NG151500.1.p 224 DDGYTWRKYGQKDILGSRYPRSYYRCTHKnyyGCEAKKKVQRLDDDPFMYEVTYCGNHTC 283
8***************************99999*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 3.4E-27 | 216 | 283 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.19E-24 | 221 | 283 | IPR003657 | WRKY domain |
| SMART | SM00774 | 6.8E-38 | 223 | 285 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 21.643 | 224 | 281 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 2.0E-25 | 224 | 283 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 417 aa Download sequence Send to blast |
MHGLAAHPGP PLALLGGSLD HRDSNSITRE LVVVAGRRAM DDAVLQIIED AFGLARELMA 60 ELQATQQNDP AYLAGRCQRI AQAYLTASRM LGPHPHGADD LSPRALPPPH HPLGDGGSGS 120 SHGHLQQLEL LRPFLGGGGA PSSAPFQQHL GRLLEPSPFN TSTTSPDMFG AGTSGGPVRR 180 QASSSRSSPP VQPRQHRMRR RESGERMTMM VPVQRTGNTD LPPDDGYTWR KYGQKDILGS 240 RYPRSYYRCT HKNYYGCEAK KKVQRLDDDP FMYEVTYCGN HTCLTSTMPL LTLPAPTTTA 300 VSSAAASMLT NSPTSSSAAA ILAGQDLVMA PAAEHPTPAL STAIQLGISW MPSTLISPSA 360 GEGSSSAQMD VPAASGTRDT EFPVMDLADA MFNSGSSGGS SMDGIFPAHH HDQRDS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6ir8_A | 2e-21 | 224 | 283 | 9 | 68 | OsWRKY45 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00304 | DAP | Transfer from AT2G40740 | Download |
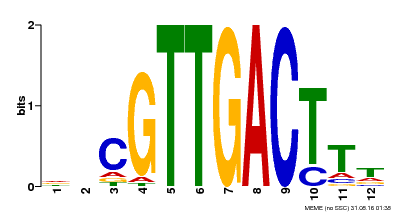
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pavir.3NG151500.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK287817 | 2e-85 | AK287817.1 Oryza sativa Japonica Group cDNA, clone: J065181J09, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025809471.1 | 0.0 | uncharacterized protein LOC112887503 | ||||
| TrEMBL | A0A2T7EFG4 | 0.0 | A0A2T7EFG4_9POAL; Uncharacterized protein | ||||
| STRING | Pavir.Ca01708.1.p | 0.0 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP9796 | 33 | 44 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G40740.1 | 2e-40 | WRKY DNA-binding protein 55 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pavir.3NG151500.1.p |



