 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pavir.4KG313000.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 227aa MW: 25992.8 Da PI: 8.8681 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 92.7 | 1.8e-29 | 9 | 59 | 1 | 51 |
S---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE- CS
SRF-TF 1 krienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklyeyss 51
krien + rqvtfskRrng+lKKA+ELSvLCdaeva++++s++gklye++s
Pavir.4KG313000.1.p 9 KRIENPTSRQVTFSKRRNGLLKKAFELSVLCDAEVALVVISPRGKLYEFAS 59
79***********************************************86 PP
| |||||||
| 2 | K-box | 71.6 | 2.4e-24 | 78 | 171 | 6 | 99 |
K-box 6 gksleeakaeslqqelakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkekelqeenkaLr 95
++++++++ e+ + +++ L k++e L+ +R+llGe+Le++s++eL++Le +Leksl iR +K++ll eq+++l++ke l+++n++Lr
Pavir.4KG313000.1.p 78 SSKTAQQDIEQVKADAEGLAKKLEALEAYKRKLLGEKLEECSIEELHSLEVKLEKSLHCIRGRKTQLLEEQVNKLKEKEMTLRKNNEELR 167
334788999********************************************************************************* PP
K-box 96 kkle 99
+k +
Pavir.4KG313000.1.p 168 EKCK 171
*976 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00432 | 7.0E-39 | 1 | 60 | IPR002100 | Transcription factor, MADS-box |
| PROSITE profile | PS50066 | 31.076 | 1 | 61 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 6.2E-30 | 3 | 23 | IPR002100 | Transcription factor, MADS-box |
| PROSITE pattern | PS00350 | 0 | 3 | 57 | IPR002100 | Transcription factor, MADS-box |
| SuperFamily | SSF55455 | 3.92E-32 | 3 | 81 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 7.43E-42 | 3 | 75 | No hit | No description |
| Pfam | PF00319 | 1.6E-26 | 10 | 57 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 6.2E-30 | 23 | 38 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 6.2E-30 | 38 | 59 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF01486 | 4.2E-25 | 82 | 171 | IPR002487 | Transcription factor, K-box |
| PROSITE profile | PS51297 | 13.642 | 86 | 176 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000060 | Biological Process | protein import into nucleus, translocation | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009911 | Biological Process | positive regulation of flower development | ||||
| GO:0010077 | Biological Process | maintenance of inflorescence meristem identity | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005737 | Cellular Component | cytoplasm | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008134 | Molecular Function | transcription factor binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 227 aa Download sequence Send to blast |
MVRGKTQMKR IENPTSRQVT FSKRRNGLLK KAFELSVLCD AEVALVVISP RGKLYEFASA 60 SVQKTIERYR TYTKDNVSSK TAQQDIEQVK ADAEGLAKKL EALEAYKRKL LGEKLEECSI 120 EELHSLEVKL EKSLHCIRGR KTQLLEEQVN KLKEKEMTLR KNNEELREKC KNQPPLPLTP 180 ARPVITVEDD RPEQNDDDRM DVETELYLGL PGRDPRRNEV AEVRSG* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5f28_A | 2e-18 | 1 | 72 | 1 | 72 | MEF2C |
| 5f28_B | 2e-18 | 1 | 72 | 1 | 72 | MEF2C |
| 5f28_C | 2e-18 | 1 | 72 | 1 | 72 | MEF2C |
| 5f28_D | 2e-18 | 1 | 72 | 1 | 72 | MEF2C |
| 6byy_A | 2e-18 | 1 | 73 | 1 | 73 | MEF2 CHIMERA |
| 6byy_B | 2e-18 | 1 | 73 | 1 | 73 | MEF2 CHIMERA |
| 6byy_C | 2e-18 | 1 | 73 | 1 | 73 | MEF2 CHIMERA |
| 6byy_D | 2e-18 | 1 | 73 | 1 | 73 | MEF2 CHIMERA |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Pvr.3488 | 0.0 | leaf| root | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Low expression in the young panicle continues to decline as the organ mature. {ECO:0000269|PubMed:15144377, ECO:0000269|PubMed:17166135}. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in mature leaves and at low levels in roots and young panicles. {ECO:0000269|PubMed:15144377, ECO:0000269|PubMed:17166135}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor active in flowering time control. May control internode elongation and promote floral transition phase. May act upstream of the floral regulators MADS1, MADS14, MADS15 and MADS18 in the floral induction pathway. {ECO:0000269|PubMed:15144377, ECO:0000269|PubMed:17166135}. | |||||
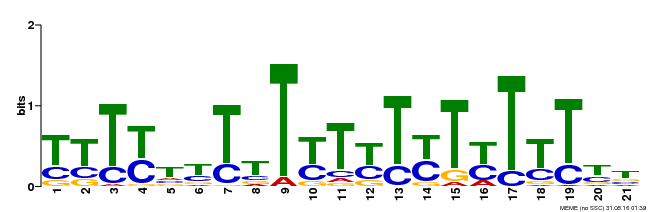
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00076 | ChIP-chip | Transfer from AT2G45660 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pavir.4KG313000.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EU960774 | 0.0 | EU960774.1 Zea mays clone 228490 MADS-box transcription factor 56 mRNA, complete cds. | |||
| GenBank | EU962504 | 0.0 | EU962504.1 Zea mays clone 243238 mRNA sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025797677.1 | 1e-150 | MADS-box transcription factor 50-like | ||||
| Refseq | XP_025797678.1 | 1e-150 | MADS-box transcription factor 50-like | ||||
| Refseq | XP_025797679.1 | 1e-150 | MADS-box transcription factor 50-like | ||||
| Swissprot | Q9XJ60 | 1e-122 | MAD50_ORYSJ; MADS-box transcription factor 50 | ||||
| TrEMBL | A0A1L5YF98 | 1e-161 | A0A1L5YF98_PANVG; SL1 | ||||
| STRING | GRMZM2G171365_P01 | 1e-125 | (Zea mays) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1413 | 33 | 79 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G45660.1 | 8e-75 | AGAMOUS-like 20 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pavir.4KG313000.1.p |



