 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pavir.5KG436500.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 321aa MW: 35095.4 Da PI: 9.0902 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 105.4 | 7e-33 | 66 | 206 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkk......leleev.ikev.diykvePwdLpkkvkaeekewyfFskrdkkyatgkrknratk... 79
lp+G++F+Ptd+el+ e+L+ k++ + ++++ i+ +i++++P++Lp v ++ +fF++ +k+y+tg+rk+r+++
Pavir.5KG436500.1.p 66 LPAGVKFDPTDQELL-EHLEGKARPDArklhplIDEFIPtIEGEnGICYTHPERLP-GVGKDGLIRHFFHRPSKAYTTGTRKRRKVHtde 153
799************.99****998885665443333333665557**********.555555677******************744456 PP
NAM 80 ...sgyWkatgkdkevlskkgelvglkktLvfy..kgrapkgektdWvmheyrl 128
+ +W++tgk+++v++ +g+l+g kk+Lv+y g+++k+ekt+Wvmh+y+l
Pavir.5KG436500.1.p 154 qggETRWHKTGKTRPVFT-NGKLKGYKKILVLYtnYGKQRKPEKTNWVMHQYHL 206
555778************.999***********6657999999*********98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 1.19E-37 | 58 | 225 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 33.262 | 66 | 225 | IPR003441 | NAC domain |
| Pfam | PF02365 | 4.2E-15 | 67 | 206 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:2000652 | Biological Process | regulation of secondary cell wall biogenesis | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 321 aa Download sequence Send to blast |
MTWCNSLNDV RAVENNLATA AAVAAAKKQQ QASSHVNLIR TCPSCGHRAQ YEQLQAAATI 60 QDLPGLPAGV KFDPTDQELL EHLEGKARPD ARKLHPLIDE FIPTIEGENG ICYTHPERLP 120 GVGKDGLIRH FFHRPSKAYT TGTRKRRKVH TDEQGGETRW HKTGKTRPVF TNGKLKGYKK 180 ILVLYTNYGK QRKPEKTNWV MHQYHLGSDE EEKDGELVVS KVFYQTQPRQ CGSGSATAKD 240 VVPLAASAAT TMDHHHHHHH DGGSGGSNSM LKEAAGIVDF YSPAALIGYN QAAPNNRAAF 300 SAHLMPNFEV HTAGAAGFGP * |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Pvr.3227 | 0.0 | root| stem | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in protoxylem and elongating interfascicular fiber cells of elongating internodes, developing metaxylem cells and interfascicular fibers of non-elongating internodes and developing secondary xylem of roots. {ECO:0000269|PubMed:18952777}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that plays a regulatory role in the development of secondary cell wall fibers (PubMed:18952777,PubMed:22133261). Involved in the regulation of cellulose and hemicellulose biosynthetic genes as well as of genes involved in lignin polymerization and signaling (PubMed:22133261). Is not a direct target of SND1 (PubMed:18952777). {ECO:0000269|PubMed:18952777, ECO:0000269|PubMed:22133261}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00459 | DAP | Transfer from AT4G28500 | Download |
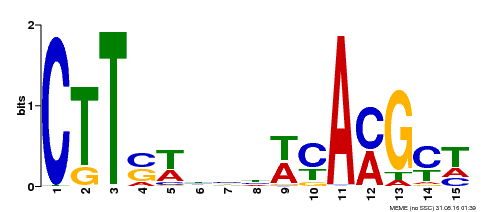
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pavir.5KG436500.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT054945 | 0.0 | BT054945.1 Zea mays full-length cDNA clone ZM_BFc0091A12 mRNA, complete cds. | |||
| GenBank | BT062976 | 0.0 | BT062976.1 Zea mays full-length cDNA clone ZM_BFc0013H24 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025814631.1 | 0.0 | NAC domain-containing protein 73-like isoform X1 | ||||
| Swissprot | O49459 | 1e-124 | NAC73_ARATH; NAC domain-containing protein 73 | ||||
| TrEMBL | A0A2T7DIP1 | 0.0 | A0A2T7DIP1_9POAL; Uncharacterized protein | ||||
| STRING | Pavir.J05241.1.p | 0.0 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3075 | 37 | 86 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G28500.1 | 1e-125 | NAC domain containing protein 73 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pavir.5KG436500.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



