 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pavir.5KG489300.2.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 264aa MW: 28444.2 Da PI: 9.1325 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 37.5 | 5.3e-12 | 5 | 56 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHT.....TTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmg.....kgRtlkqcksrwqky 47
+++WT eE+e l ++v ++G g W+tI + R+ ++k++w+++
Pavir.5KG489300.2.p 5 KQKWTSEEEEALRRGVLKHGAGKWRTIQKDPEfspvlSSRSNIDLKDKWRNL 56
79***********************************9************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 13.143 | 1 | 60 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.22E-15 | 2 | 58 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.2E-7 | 4 | 59 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.3E-8 | 5 | 56 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.9E-14 | 5 | 56 | IPR009057 | Homeodomain-like |
| CDD | cd11660 | 6.71E-21 | 6 | 56 | No hit | No description |
| Gene3D | G3DSA:1.10.10.10 | 3.4E-6 | 115 | 159 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SuperFamily | SSF46785 | 8.5E-6 | 115 | 159 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006334 | Biological Process | nucleosome assembly | ||||
| GO:0000786 | Cellular Component | nucleosome | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 264 aa Download sequence Send to blast |
MGAPKQKWTS EEEEALRRGV LKHGAGKWRT IQKDPEFSPV LSSRSNIDLK DKWRNLSFSA 60 SGMGSRDKTR GPRTTGPSCS PLSTSQASSQ ASVAPVANKV AEAPPGAEKK QQEVKPHPKY 120 GAMILEALSE LNEPNGTELT EICSFIEVDT KSYRLADSFA TRTPAPIKAS APKQKDPSKP 180 SKVSKNLGLF ATSSPALEAA MAAAAKVADA EVKAHDAHDQ MMEAERILKM AEKTESLLTI 240 AAEIYDRCSR GEITTLNPPQ REV* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Pvr.11511 | 1e-110 | callus| leaf| stem | ||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds preferentially double-stranded telomeric repeats, but may also bind to the single telomeric strand. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00231 | DAP | Transfer from AT1G72740 | Download |
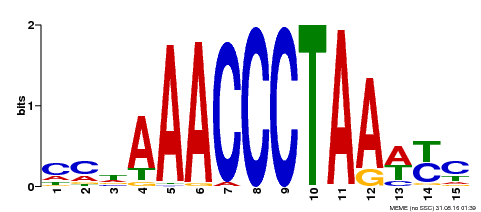
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pavir.5KG489300.2.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025815372.1 | 1e-140 | glycylpeptide N-tetradecanoyltransferase 1 | ||||
| Swissprot | Q6WLH4 | 1e-113 | SMH3_MAIZE; Single myb histone 3 | ||||
| TrEMBL | A0A2S3HSG5 | 1e-153 | A0A2S3HSG5_9POAL; Uncharacterized protein | ||||
| TrEMBL | A0A2T7DHT0 | 1e-153 | A0A2T7DHT0_9POAL; Uncharacterized protein | ||||
| STRING | Pavir.Ea02570.1.p | 1e-167 | (Panicum virgatum) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G49950.3 | 2e-34 | telomere repeat binding factor 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pavir.5KG489300.2.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



