 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pavir.5KG710000.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | B3 | ||||||||
| Protein Properties | Length: 703aa MW: 74230.5 Da PI: 6.5716 | ||||||||
| Description | B3 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 61.2 | 1.7e-19 | 533 | 629 | 2 | 95 |
EEE-..-HHHHTT-EE--HHH.HTT..---..--SEEEEEE.TTS-EEEEEE....EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-S CS
B3 2 fkvltpsdvlksgrlvlpkkfaeeh..ggkkeesktltled.esgrsWevkliy..rkksgryvltkGWkeFvkangLkegDfvvFkldg 86
+kvl++sdv+++gr+vlpkk ae h ++k++ ++ + +ed ++r+W++++++ ++ks++y+l+ ++ eFv++n+L+egDf+v++ +
Pavir.5KG710000.1.p 533 QKVLKQSDVGSLGRIVLPKKEAEVHlpELKTRDGISIPMEDiGTSRVWNMRYRFwpNNKSRMYLLE-NTGEFVRSNELQEGDFIVIY--S 619
79*************************999***********7778*******99777777777777.********************..5 PP
S.SEE..EEE CS
B3 87 r.sefelvvk 95
+ ++++++++
Pavir.5KG710000.1.p 620 DvKSGKYLIR 629
5588877776 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.40.330.10 | 2.3E-27 | 526 | 637 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10015 | 1.00E-31 | 530 | 631 | No hit | No description |
| SuperFamily | SSF101936 | 3.14E-22 | 531 | 627 | IPR015300 | DNA-binding pseudobarrel domain |
| PROSITE profile | PS50863 | 10.532 | 532 | 634 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 1.3E-18 | 532 | 634 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 6.2E-16 | 533 | 628 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009657 | Biological Process | plastid organization | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009793 | Biological Process | embryo development ending in seed dormancy | ||||
| GO:0031930 | Biological Process | mitochondria-nucleus signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005829 | Cellular Component | cytosol | ||||
| GO:0001076 | Molecular Function | transcription factor activity, RNA polymerase II transcription factor binding | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 703 aa Download sequence Send to blast |
MDASAVSSPP HSQENPPEHG GALGEAPAEE IGGEAADDFL FAEDTFPSLP DFPCLSSPSS 60 STFSSSSSSN SSCAYTNAAG GAGGGGADGA AGDRSVPASA GEGFDALDDI DQLLDFASLS 120 MPWDSEPTFP EVSMMLEDAM FAPPHPVGDG TREGKAVLEG TGGEEACMDA GAAGEDLPRF 180 FMEWLTSNRE NISAEDLRSI RLRRSTIEAA AARLGGGRQG TMQLLKLILT WVQNHHLQRK 240 RPRDALEEAA GLHGHGHSQL SSPGGNPGYE FPAGVQDMTA GGGTSWMPYQ QPFTPPACGG 300 DAVYQSAAGQ YPFHQSSSTS SVVVSSQPFS PPAVGDMHAA GGGNMAWTQP YVPYPGASMG 360 SYPLPPVVPQ PFSSGFGGQY AGAGHPMAPQ RMAAVEASAT KEARKKRMAR QRRLSCLQQQ 420 RSQQLNLGQI QVPVLQQEPS ARSTNSAPVT PAAGGWGFWS PGSPQQVQNA LPKSNSSRAP 480 MQQVPRSPES APPPPPPAKP APGARQEESP QRSMAADKRQ GAKTDKNLRY LLQKVLKQSD 540 VGSLGRIVLP KKEAEVHLPE LKTRDGISIP MEDIGTSRVW NMRYRFWPNN KSRMYLLENT 600 GEFVRSNELQ EGDFIVIYSD VKSGKYLIRG VKVRPAQEQQ GNGSSSLGKH KHGYPGPEKA 660 GGAPDDGGAD GTNKPDGGCK GRSPQGVRRA RHQGAASMAV SI* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j9b_A | 6e-37 | 528 | 638 | 1 | 111 | B3 domain-containing transcription factor FUS3 |
| 6j9b_D | 6e-37 | 528 | 638 | 1 | 111 | B3 domain-containing transcription factor FUS3 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Pvr.4716 | 0.0 | callus | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Between 10 and 30 days after pollination. | |||||
| Uniprot | TISSUE SPECIFICITY: Seed. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator specifically required for expression of the maturation program in the seed development. Probably potentiates the response to the seed-specific hormone abscisic acid (ABA). May bind to DNA indirectly. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00083 | PBM | Transfer from AT3G24650 | Download |
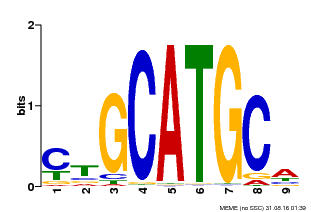
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pavir.5KG710000.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT060994 | 0.0 | BT060994.1 Zea mays full-length cDNA clone ZM_BFb0082K18 mRNA, complete cds. | |||
| GenBank | JX469991 | 0.0 | JX469991.1 Zea mays subsp. mays clone UT3351 ABI3VP1-type transcription factor mRNA, partial cds. | |||
| GenBank | MZEREGPRO | 0.0 | M60214.1 Zea mays viviparous-1 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025816909.1 | 0.0 | regulatory protein viviparous-1 | ||||
| Swissprot | P26307 | 0.0 | VIV1_MAIZE; Regulatory protein viviparous-1 | ||||
| TrEMBL | A0A2T8IIY4 | 0.0 | A0A2T8IIY4_9POAL; Uncharacterized protein | ||||
| STRING | Pavir.Ea03476.1.p | 0.0 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP7529 | 34 | 43 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G24650.1 | 8e-67 | B3 family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pavir.5KG710000.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



