 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pavir.5NG568500.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 559aa MW: 59644.2 Da PI: 7.6667 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 105.2 | 3.5e-33 | 221 | 277 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
dDgynWrKYGqK+vkgse+prsYY+Ct+ gCp+kkkvers d++++ei+Y+g+Hnh
Pavir.5NG568500.1.p 221 DDGYNWRKYGQKQVKGSENPRSYYKCTFLGCPTKKKVERSL-DGQITEIVYKGTHNHA 277
8****************************************.***************7 PP
| |||||||
| 2 | WRKY | 105.9 | 2.1e-33 | 383 | 441 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+vkg+++prsYY+Ct+agCpv+k+ver+++d ++v++tYeg+Hnh+
Pavir.5NG568500.1.p 383 LDDGYRWRKYGQKVVKGNPNPRSYYKCTTAGCPVRKHVERASHDLRAVITTYEGKHNHD 441
59********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 8.0E-29 | 212 | 279 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 24.157 | 215 | 279 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 3.14E-25 | 216 | 279 | IPR003657 | WRKY domain |
| SMART | SM00774 | 2.8E-36 | 220 | 278 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 2.6E-25 | 221 | 276 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 2.7E-37 | 368 | 443 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.96E-29 | 375 | 443 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 38.687 | 378 | 443 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.2E-38 | 383 | 442 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 9.3E-26 | 384 | 441 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0009788 | Biological Process | negative regulation of abscisic acid-activated signaling pathway | ||||
| GO:0009938 | Biological Process | negative regulation of gibberellic acid mediated signaling pathway | ||||
| GO:0010120 | Biological Process | camalexin biosynthetic process | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0010508 | Biological Process | positive regulation of autophagy | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0070370 | Biological Process | cellular heat acclimation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 559 aa Download sequence Send to blast |
MTTSSSGSIE APANSRPGSF SFASTNFTVM LGGSAAAAGG ASRYKAMTPP SLPVSPPPVS 60 PSSFFNIPGG LNPADFLDSP ALLTSSFFPS PTTNAFASQQ FSWLTPPGAE QGGKDEQRQS 120 YPDFSFQTAP TTEEAVRTTT TFQQPPVPPA PLGEEAYRSQ QQQQQQPWSY HQQQPGMDAG 180 SSQAAYGGQF QAGSSDAAAM APHAPASGGY SQAQSQRRSS DDGYNWRKYG QKQVKGSENP 240 RSYYKCTFLG CPTKKKVERS LDGQITEIVY KGTHNHAKPQ NTRRNSSAAA QLLQGAGEAS 300 EHSFGGTPVA TPENSSASFG DDEAAGVGSP RAGNAGGDEF DEDEPDSKRW RKDGDGEGIS 360 MAGNRTVREP RVVVQTMSDI DILDDGYRWR KYGQKVVKGN PNPRSYYKCT TAGCPVRKHV 420 ERASHDLRAV ITTYEGKHNH DVPAARGSAA LYRPAPPPTD TYLAAAPGVR PPAPSMAYQT 480 GQRYGFGGQS SFGLGAGAPA QQSGGGGFGF PSGFDNPMGS YMSQHQQQQR QNDAMHASRA 540 KEEPREDMAF FPQSMMYN* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 4e-36 | 212 | 444 | 8 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 4e-36 | 212 | 444 | 8 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Pvr.431 | 8e-70 | root| stem | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: In dry seeds, expressed in aleurone cells and embryos. Levels drop rapidly but transiently in the embryos of imbibed seeds. {ECO:0000250|UniProtKB:Q6IEQ7}. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in aleurone cells. Mostly expressed in aleurone layers and leaves, and, to a lower extent, in roots, panicles and embryos. {ECO:0000250|UniProtKB:Q6IEQ7}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription repressor (By similarity). Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element. Negative regulator of both gibberellic acid (GA) and abscisic acid (ABA) signaling in aleurone cells, probably by interfering with GAM1, via the specific repression of GA- and ABA-induced promoters (By similarity). {ECO:0000250|UniProtKB:Q6IEQ7, ECO:0000250|UniProtKB:Q6QHD1}. | |||||
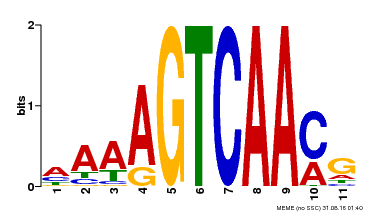
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00299 | DAP | Transfer from AT2G38470 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pavir.5NG568500.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by abscisic acid (ABA) in aleurone cells, embryos, roots and leaves (PubMed:25110688). Slightly down-regulated by gibberellic acid (GA) (By similarity). Accumulates in response to jasmonic acid (MeJA) (PubMed:16919842). {ECO:0000250|UniProtKB:Q6IEQ7, ECO:0000269|PubMed:16919842, ECO:0000269|PubMed:25110688}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT053994 | 0.0 | BT053994.1 Zea mays full-length cDNA clone ZM_BFc0103O21 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004970421.3 | 0.0 | WRKY transcription factor WRKY24 | ||||
| Swissprot | Q6B6R4 | 0.0 | WRK24_ORYSI; WRKY transcription factor WRKY24 | ||||
| TrEMBL | A0A1E5WCJ0 | 0.0 | A0A1E5WCJ0_9POAL; Putative WRKY transcription factor 33 | ||||
| STRING | Pavir.Eb03590.1.p | 0.0 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2992 | 36 | 87 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G38470.1 | 7e-96 | WRKY DNA-binding protein 33 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pavir.5NG568500.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



