 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pavir.6KG070500.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 721aa MW: 78993.7 Da PI: 6.5195 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 47.8 | 3.2e-15 | 24 | 68 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r rWT+ E++++++a k++G W +I +++g ++t+ q++s+ qk+
Pavir.6KG070500.1.p 24 RERWTEAEHKRFLEALKLYGRA-WQRIEEHVG-TKTAVQIRSHAQKF 68
78******************88.*********.************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 2.84E-16 | 18 | 74 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 19.147 | 19 | 73 | IPR017930 | Myb domain |
| TIGRFAMs | TIGR01557 | 1.6E-16 | 22 | 71 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 6.5E-12 | 23 | 71 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.5E-12 | 24 | 67 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 9.0E-8 | 24 | 64 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.03E-8 | 26 | 69 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 721 aa Download sequence Send to blast |
MEVNSSGEEM VVKVRKPYTI TKQRERWTEA EHKRFLEALK LYGRAWQRIE EHVGTKTAVQ 60 IRSHAQKFFT KLEKEAMNNG TSPGQAHDID IPPPRPKRKP NSPYPRKSGL SSETPSKEVP 120 NDKSTKSNIT VGNGNVQMAG DASLQKFQRK EVSEKGSCSE VLNLFRDAPS ASFSSVNKSS 180 SNNGVLRGIE LTKKEIRDMT TMEKNSINPT MQEDVKEIND QEMGRVNGIH VSSKCDHSNE 240 EYLDFSMQQM KLHPKSLETT YVDKQTARTS HTLAERNGAT SIPVTGTEGT HPDQTSDQVG 300 VNGSMNPCIH PMLSMDLKFD SSATPQSFPH NYAAFAPVMQ CNCNQDTYRS FVNMSSTFSS 360 MLVSTLLSNP AVHAAARLAA SYWPAAEGNT HVDPNQENPA EGVQGRNLGS PPSMASIVAA 420 TVAAASAWWA TQGLLPFLAP PMAFPFVPAP SAAFPTADLP RPSEKDRDCL VENAQKECQE 480 AQKQGQSEAL RVVASSESGG SRKGDMSIHT ELKISHVLNA EATPTTGADT SDALRNKKKQ 540 DRSSCGSNTP SSSDVEAENV PEKEDKANDK AKQASCSNSS AGDTNHRRFR SSGSTSDSWK 600 EVSEEGRLAF DALFSREKLP QSFSPPQAEE SKEVANEEED EVTTVTVDLN KNVTSIDHEL 660 DMMDEPRTSF PNELSHLKLK SRKTGFKPYK RCSVEAKENR APASDEVGTK RIRLESEAST 720 * |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Pvr.1937 | 0.0 | callus| leaf| root| stem | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00654 | PBM | Transfer from LOC_Os08g06110 | Download |
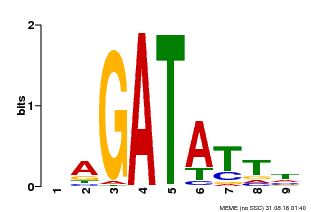
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pavir.6KG070500.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EU955544 | 0.0 | EU955544.1 Zea mays clone 1536169 LHY protein mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025820794.1 | 0.0 | protein LHY isoform X1 | ||||
| Refseq | XP_025820795.1 | 0.0 | protein LHY isoform X1 | ||||
| TrEMBL | A0A2T8IF63 | 0.0 | A0A2T8IF63_9POAL; Uncharacterized protein | ||||
| STRING | Pavir.Fa01905.1.p | 0.0 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6031 | 34 | 47 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G46830.1 | 2e-41 | circadian clock associated 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pavir.6KG070500.1.p |



