 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pavir.6NG206000.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | Dof | ||||||||
| Protein Properties | Length: 406aa MW: 41262.8 Da PI: 9.2802 | ||||||||
| Description | Dof family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-Dof | 123.7 | 6.2e-39 | 113 | 173 | 2 | 62 |
zf-Dof 2 kekalkcprCdstntkfCyynnyslsqPryfCkaCrryWtkGGalrnvPvGggrrknkkss 62
+e lkcprC+stntkfCy+nnyslsqPr+fCkaCrryWt+GGalrnvPvGgg+r+nk+s+
Pavir.6NG206000.1.p 113 PEPGLKCPRCESTNTKFCYFNNYSLSQPRHFCKACRRYWTRGGALRNVPVGGGCRRNKRSK 173
56789*****************************************************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| ProDom | PD007478 | 3.0E-36 | 101 | 171 | IPR003851 | Zinc finger, Dof-type |
| Pfam | PF02701 | 1.5E-32 | 117 | 171 | IPR003851 | Zinc finger, Dof-type |
| PROSITE profile | PS50884 | 29.646 | 117 | 171 | IPR003851 | Zinc finger, Dof-type |
| PROSITE pattern | PS01361 | 0 | 119 | 155 | IPR003851 | Zinc finger, Dof-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 406 aa Download sequence Send to blast |
MVFSSVPVYL DPPNWNQHQQ QQQQAHHGQL PSGGGSGGGV EAGHAHHHQQ HHQLPPVPPP 60 GALMAPRPDM ATIVIAPSGG GGCGAGGGGP TGGSSVRPGS MTERARLAKI PQPEPGLKCP 120 RCESTNTKFC YFNNYSLSQP RHFCKACRRY WTRGGALRNV PVGGGCRRNK RSKSSKSSSS 180 SAASASATGG TSSSTSSTTT GGSSAAAAIM PPQGQLPFMA SLHHHLGGDH YSTGASRLGF 240 PGLSSQLDPV DYQLGAGTAI GLEQWRLPQI QQQFHFLSRP DAVPPPMSGI YPFDVEGHGD 300 ATAGGFAGQM LGGSSKVPGS AGLITQLASV KMEDNPPSTA MTSLSSPREF LGLPGNLQFW 360 GGGNGGASGN NGGTANAGGG GANTAPGSSW VDLSGFNSPS SGNIL* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Pvr.20062 | 4e-71 | stem | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00295 | DAP | Transfer from AT2G37590 | Download |
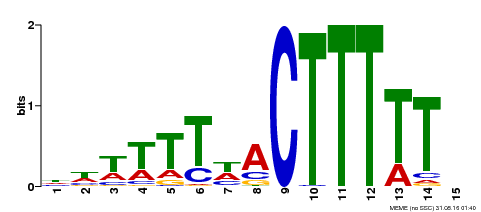
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pavir.6NG206000.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025792577.1 | 1e-169 | dof zinc finger protein DOF2.4-like | ||||
| TrEMBL | A0A3L6SDN7 | 1e-173 | A0A3L6SDN7_PANMI; Dof zinc finger protein DOF2.4-like | ||||
| STRING | Pavir.J14743.1.p | 0.0 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP4893 | 29 | 61 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G55370.2 | 2e-41 | OBF-binding protein 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pavir.6NG206000.1.p |



