 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pavir.6NG313700.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 342aa MW: 36776.9 Da PI: 7.565 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 169.3 | 1.2e-52 | 28 | 155 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpkkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdk 90
lppGfrFhPtdeelv++yL++kv + + i+evd++++ePw+Lp +++ +e+ewyfFs rd+ky+tg r+nrat +gyWkatgkd+
Pavir.6NG313700.1.p 28 LPPGFRFHPTDEELVTFYLAAKVLNGACCG-VDIAEVDLNRCEPWELPDAARMGEREWYFFSLRDRKYPTGMRTNRATGAGYWKATGKDR 116
79************************9777.34***************8888899*********************************** PP
NAM 91 evlsk.kgelvglkktLvfykgrapkgektdWvmheyrl 128
evl++ +g+l g+kktLvfykgrap+gekt+Wv+heyrl
Pavir.6NG313700.1.p 117 EVLNAaTGALLGMKKTLVFYKGRAPRGEKTKWVLHEYRL 155
***9878899***************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 4.18E-63 | 17 | 178 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 58.18 | 28 | 178 | IPR003441 | NAC domain |
| Pfam | PF02365 | 1.9E-27 | 29 | 155 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010014 | Biological Process | meristem initiation | ||||
| GO:0010199 | Biological Process | organ boundary specification between lateral organs and the meristem | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 342 aa Download sequence Send to blast |
MHHQAMVGDA LWDLLGEEMA AAAGEHGLPP GFRFHPTDEE LVTFYLAAKV LNGACCGVDI 60 AEVDLNRCEP WELPDAARMG EREWYFFSLR DRKYPTGMRT NRATGAGYWK ATGKDREVLN 120 AATGALLGMK KTLVFYKGRA PRGEKTKWVL HEYRLDGDFA AARRSCKEEW VICRILHKAG 180 DQYSKLMMVK NPYYLPMGVD PSSFCFQQDP AAPALPNPSG FSTAGLSFHH GHHPGMHPPP 240 LPANQLKLSN GRGFPASACT QEPPPSGCGG SSNAAVGMPP YPPPFTSIVA GKPAAPPPQA 300 PGVVNAGAQE PPAPSPTWLE AYVQHGGFLY EMGPAAAPRG A* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 2e-48 | 28 | 178 | 17 | 165 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 2e-48 | 28 | 178 | 17 | 165 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 2e-48 | 28 | 178 | 17 | 165 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 2e-48 | 28 | 178 | 17 | 165 | NO APICAL MERISTEM PROTEIN |
| 3swm_A | 2e-48 | 15 | 178 | 7 | 168 | NAC domain-containing protein 19 |
| 3swm_B | 2e-48 | 15 | 178 | 7 | 168 | NAC domain-containing protein 19 |
| 3swm_C | 2e-48 | 15 | 178 | 7 | 168 | NAC domain-containing protein 19 |
| 3swm_D | 2e-48 | 15 | 178 | 7 | 168 | NAC domain-containing protein 19 |
| 3swp_A | 2e-48 | 15 | 178 | 7 | 168 | NAC domain-containing protein 19 |
| 3swp_B | 2e-48 | 15 | 178 | 7 | 168 | NAC domain-containing protein 19 |
| 3swp_C | 2e-48 | 15 | 178 | 7 | 168 | NAC domain-containing protein 19 |
| 3swp_D | 2e-48 | 15 | 178 | 7 | 168 | NAC domain-containing protein 19 |
| 4dul_A | 2e-48 | 28 | 178 | 17 | 165 | NAC domain-containing protein 19 |
| 4dul_B | 2e-48 | 28 | 178 | 17 | 165 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Pvr.18171 | 0.0 | stem | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: First expressed at the globular stage, mostly in the apical part of the embryo. During the triangular stage, confined to the boundary of emerging cotyledons. Later restricted to the center of the apical part of the embryo and to seedling apex, at the boundaries of the cotyledon margins and the boundaries between the SAM and the cotyledons. Localized in a one-cell-wide ring at the boundary between trichomes or lateral roots and epidermis. Accumulates at the boundaries between leaf primordia and the shoot meristem and between floral primordia and the inflorescence meristem. Found in the adaxial axils of secondary inflorescences and pedicels, and in axiallary buds. In flowers, expressed in a ring at the bases of sepals and petals. In carpels, confined to the boundaries between ovule primordia. In ovules, localized in a ring at the boundary between the nucellus and the chalaza. In the mature embryo sac, detected in the two polar nuclei of the central cell. {ECO:0000269|PubMed:12837947, ECO:0000269|PubMed:17122068}. | |||||
| Uniprot | TISSUE SPECIFICITY: In a general manner, present at the boundaries between mersitems and araising primordia. {ECO:0000269|PubMed:12837947}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator. Involved in molecular mechanisms regulating shoot apical meristem (SAM) formation during embryogenesis and organ separation. Required for axillary meristem initiation and separation of the meristem from the main stem. May act as an inhibitor of cell division. {ECO:0000269|PubMed:12837947, ECO:0000269|PubMed:17122068}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00241 | DAP | Transfer from AT1G76420 | Download |
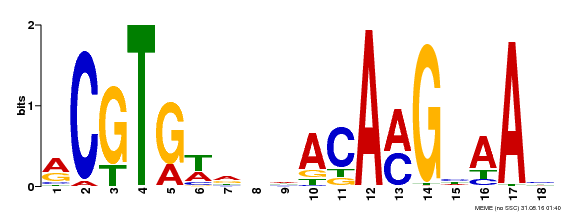
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pavir.6NG313700.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By BRM, at the chromatin level, and conferring a very specific spatial expression pattern. {ECO:0000269|PubMed:16854978}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT061270 | 0.0 | BT061270.1 Zea mays full-length cDNA clone ZM_BFb0128J16 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025822038.1 | 0.0 | protein CUP-SHAPED COTYLEDON 3-like | ||||
| Swissprot | Q9S851 | 2e-88 | NAC31_ARATH; Protein CUP-SHAPED COTYLEDON 3 | ||||
| TrEMBL | A0A2T7D9C0 | 0.0 | A0A2T7D9C0_9POAL; Uncharacterized protein | ||||
| STRING | Pavir.Fb01938.1.p | 0.0 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP11237 | 33 | 41 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G76420.1 | 2e-86 | NAC family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pavir.6NG313700.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



