 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pavir.7KG061000.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 338aa MW: 35967.5 Da PI: 10.2485 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 98.6 | 4.1e-31 | 260 | 318 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-ST.T---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsa.gCpvkkkversaedpkvveitYegeHnhe 59
D+y+WrKYGqK++kgs++pr+YY+C++ gCp++k+ver+ dp+++++tYegeH+h+
Pavir.7KG061000.1.p 260 ADEYSWRKYGQKPIKGSPYPRGYYKCSTVrGCPARKHVERDPGDPSMLIVTYEGEHRHS 318
69************************9888****************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF10533 | 2.2E-19 | 208 | 257 | IPR018872 | Zn-cluster domain |
| Gene3D | G3DSA:2.20.25.80 | 2.3E-32 | 248 | 318 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 30.458 | 254 | 320 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 3.53E-26 | 257 | 319 | IPR003657 | WRKY domain |
| SMART | SM00774 | 3.9E-37 | 259 | 319 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 3.3E-26 | 261 | 317 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009938 | Biological Process | negative regulation of gibberellic acid mediated signaling pathway | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:1990841 | Molecular Function | promoter-specific chromatin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 338 aa Download sequence Send to blast |
MMTLDLMGRY GRVDEKVAIQ EAAAAGLRGM EHLISQLSRA GTGTGERSSS SPGAAPAQNQ 60 QQPEQSHLQQ QQQQLLLLQE VDCREITDMT VSKFKKVISI LNRTGHARFR RGPVVAQSPG 120 PAEHAARSAP APRPVTLDFT KSVSGYSRDS GFSVSGASSS FLSSVTTGDG SVSNGRGGGG 180 SSSLMLLPPA GAASCGKPPL SSSGAGQKRR CHDHAHSENV AGGKYGATGG RCHCSKRRKH 240 RVKRTIRVPA ISPKVSDIPA DEYSWRKYGQ KPIKGSPYPR GYYKCSTVRG CPARKHVERD 300 PGDPSMLIVT YEGEHRHSPE DPPAPPPLGP LPELPNH* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2ayd_A | 2e-20 | 247 | 317 | 2 | 71 | WRKY transcription factor 1 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Pvr.14390 | 1e-119 | root | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Highly expressed in aleurone cells. In seeds, predominantly present in the plumule, radicle and scutellum of the embryo. {ECO:0000250|UniProtKB:Q0JEE2}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Interacts, when in complex with WRKY71, specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element. Represses specifically gibberellic acid (GA)-induced promoters in aleurone cells, probably by interfering with GAM1. {ECO:0000250|UniProtKB:Q0JEE2}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00464 | DAP | Transfer from AT4G31550 | Download |
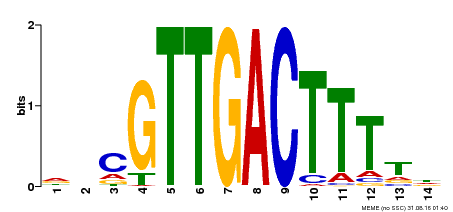
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pavir.7KG061000.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by abscisic acid (ABA) in aleurone cells (By similarity) (PubMed:15618416). Accumulates in response to uniconazole, a gibberellic acid (GA) biosynthesis inhibitor (By similarity). Repressed by GA (PubMed:15618416). {ECO:0000250|UniProtKB:Q0JEE2, ECO:0000269|PubMed:15618416}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT042579 | 0.0 | BT042579.1 Zea mays full-length cDNA clone ZM_BFb0345E21 mRNA, complete cds. | |||
| GenBank | EU966905 | 0.0 | EU966905.1 Zea mays clone 298088 WRKY51 - superfamily of TFs having WRKY and zinc finger domains mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025826098.1 | 0.0 | WRKY transcription factor WRKY51-like | ||||
| Swissprot | Q6IEN1 | 1e-143 | WRK51_ORYSI; WRKY transcription factor WRKY51 | ||||
| TrEMBL | A0A3L6QBB2 | 0.0 | A0A3L6QBB2_PANMI; Putative WRKY transcription factor 17 | ||||
| STRING | Pavir.Ga02680.1.p | 0.0 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP617 | 38 | 171 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G31550.1 | 3e-88 | WRKY DNA-binding protein 11 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pavir.7KG061000.1.p |



