 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pavir.7NG225600.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 384aa MW: 41725.4 Da PI: 6.5241 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 82.8 | 4.5e-26 | 45 | 114 | 2 | 71 |
trihelix 2 WtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkr 71
W ++e+l+Li++rrem+ +++++k++k+lWe++s++mr++gf rsp++C++kw+n+ k++kk + ++++
Pavir.7NG225600.1.p 45 WVREETLCLIALRREMDAHFNTSKSNKHLWEAISARMRDQGFDRSPTMCTDKWRNVLKEFKKARSHARSS 114
****************************************************************998874 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50090 | 8.432 | 37 | 101 | IPR017877 | Myb-like domain |
| Gene3D | G3DSA:1.10.10.60 | 1.2E-4 | 38 | 103 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.3E-5 | 41 | 103 | IPR001005 | SANT/Myb domain |
| CDD | cd12203 | 4.57E-27 | 43 | 108 | No hit | No description |
| Pfam | PF13837 | 8.0E-18 | 44 | 135 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 384 aa Download sequence Send to blast |
MLLSGPASAP TPPLLLPESS GEDCVHDSSR AAAAGSAPKR RAETWVREET LCLIALRREM 60 DAHFNTSKSN KHLWEAISAR MRDQGFDRSP TMCTDKWRNV LKEFKKARSH ARSSGGGGAG 120 GNANAKMAYY KEIDDLLKRR GKASGGGGGG GCVGTGSGSG AGKSPTSNSK IESYLQFTTD 180 NGFEDANIPF GPVEANGTSI LSIDDRLEDD RHPLPLAAAD AVATNGVNPW NWRDTSTNGG 240 DNQGTFGGRV ILVKWGDYTK RIGIDGTSEA IKAAIKSAFG LRTRRALWLE DEDEIVRTLD 300 RDMPIGTYTL HLDDGNCVTI KLCDGSRMQT SEDKTFYTEE DFRDFLTRRG WTLLREYGGY 360 RNVDSLDDLR PGVIYQGLRS LGD* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2jmw_A | 1e-37 | 39 | 132 | 1 | 86 | DNA binding protein GT-1 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Pvr.1811 | 0.0 | callus| leaf| stem | ||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
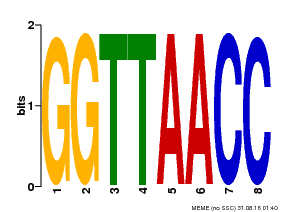
| UniProt | Probable transcription factor that binds specifically to the core DNA sequence 5'-GGTTAA-3'. May act as a molecular switch in response to light signals. {ECO:0000269|PubMed:10437822, ECO:0000269|PubMed:15044016, ECO:0000269|PubMed:7866025}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00004 | PBM | Transfer from 920224 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pavir.7NG225600.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001146488.1 | 0.0 | uncharacterized LOC100280076 | ||||
| Swissprot | Q9FX53 | 1e-138 | TGT1_ARATH; Trihelix transcription factor GT-1 | ||||
| TrEMBL | A0A3L6GEX5 | 0.0 | A0A3L6GEX5_MAIZE; Trihelix transcription factor GT-1 | ||||
| STRING | Pavir.Gb01230.1.p | 0.0 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP5631 | 37 | 52 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G13450.1 | 1e-127 | Trihelix family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pavir.7NG225600.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



