 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pavir.9NG072500.3.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | TALE | ||||||||
| Protein Properties | Length: 274aa MW: 30817.9 Da PI: 6.3173 | ||||||||
| Description | TALE family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 29.1 | 1.7e-09 | 195 | 230 | 20 | 55 |
HHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHH CS
Homeobox 20 eknrypsaeereeLAkklgLterqVkvWFqNrRake 55
+k +yps++e+ LA+ +gL+++q+ +WF N+R ++
Pavir.9NG072500.3.p 195 YKWPYPSETEKLALAETTGLDQKQINNWFINQRKRR 230
4679***************************99763 PP
| |||||||
| 2 | ELK | 43.3 | 7.2e-15 | 150 | 171 | 1 | 22 |
ELK 1 ELKhqLlrKYsgyLgsLkqEFs 22
ELKhqLlrKY+gyLg+L+qEFs
Pavir.9NG072500.3.p 150 ELKHQLLRKYGGYLGGLRQEFS 171
9********************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01255 | 3.3E-21 | 17 | 61 | IPR005540 | KNOX1 |
| Pfam | PF03790 | 2.5E-22 | 18 | 59 | IPR005540 | KNOX1 |
| SMART | SM01256 | 2.0E-27 | 69 | 120 | IPR005541 | KNOX2 |
| Pfam | PF03791 | 3.2E-23 | 74 | 119 | IPR005541 | KNOX2 |
| SMART | SM01188 | 4.2E-8 | 150 | 171 | IPR005539 | ELK domain |
| PROSITE profile | PS51213 | 11.575 | 150 | 170 | IPR005539 | ELK domain |
| Pfam | PF03789 | 2.7E-11 | 150 | 171 | IPR005539 | ELK domain |
| PROSITE profile | PS50071 | 12.957 | 170 | 233 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 5.99E-19 | 171 | 242 | IPR009057 | Homeodomain-like |
| SMART | SM00389 | 1.5E-10 | 172 | 237 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 1.9E-26 | 175 | 236 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 2.37E-11 | 182 | 233 | No hit | No description |
| Pfam | PF05920 | 1.5E-16 | 190 | 229 | IPR008422 | Homeobox KN domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 274 aa Download sequence Send to blast |
MAGAVLQREI SPVDHAETIK AKIMSHPQYS ALVAAYLDCQ KVGAPPDVSD RLSAMAAKLD 60 AQPGPSWRRH EPARADPELD QFMEAYCNML VKYQEELARP IHEAAEFFKS VERQLDSITD 120 SNYCEGAGSS EDDQDASCPE EIDPCAEDKE LKHQLLRKYG GYLGGLRQEF SKRKKKGKLP 180 KEARQKLLHW WELHYKWPYP SETEKLALAE TTGLDQKQIN NWFINQRKRR WKPTSEDMPF 240 AMVEAAGGFH APQGTAAAAA VLYMADGMYR LGS* |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Expressed in the embryo at 4 days after pollination (DAP) in the ventral and basal part of the embryo including the initial of the shoot apical meristem (SAM). At 5 DAP, expressed at the ventral side of the embryo, but the expression within SAM is down-regulated. At 7 DAP, expression is restricted to the boundaries between the embryonic organs, between the scutellum and the coleoptile, the coleoptile and the second leaf primordium, the shoot apical meristem and the first leaf primordium, the first leaf primordium and the coleoptile, the coleoptile and the epiblast and at the tip of the epiblast, but not in the leaf primordia. {ECO:0000269|PubMed:10080693}. | |||||
| Uniprot | DEVELOPMENTAL STAGE: Expressed in the embryo at 4 days after pollination (DAP) in the ventral and basal part of the embryo, including the initial of the shoot apical meristem (SAM). At 5 DAP, expressed at the ventral side of the embryo, but the expression within SAM is down-regulated. At 7 DAP, expression is restricted to the boundaries between the embryonic organs, between the scutellum and the coleoptile, the coleoptile and the second leaf primordium, the shoot apical meristem and the first leaf primordium, the first leaf primordium and the coleoptile, the coleoptile and the epiblast and at the tip of the epiblast, but not in the leaf primordia. {ECO:0000269|PubMed:10080693, ECO:0000269|PubMed:10095070, ECO:0000269|PubMed:9869405}. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in stems, rachis and inflorescence. {ECO:0000269|PubMed:9869405}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may be involved in shoot formation during embryogenesis. {ECO:0000269|PubMed:10080693}. | |||||
| UniProt | Probable transcription factor that may be involved in shoot formation during embryogenesis. {ECO:0000269|PubMed:10080693, ECO:0000269|PubMed:9869405}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00671 | PBM | Transfer from GRMZM2G135447 | Download |
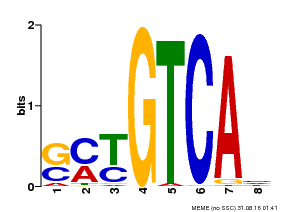
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pavir.9NG072500.3.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EU966373 | 0.0 | EU966373.1 Zea mays clone 293957 homeobox protein rough sheath 1 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025795146.1 | 1e-174 | homeobox protein knotted-1-like 12 isoform X3 | ||||
| Swissprot | O65034 | 1e-114 | KNOSC_ORYSI; Homeobox protein knotted-1-like 12 | ||||
| Swissprot | O80416 | 1e-114 | KNOSC_ORYSJ; Homeobox protein knotted-1-like 12 | ||||
| TrEMBL | A0A2T7C0H0 | 1e-175 | A0A2T7C0H0_9POAL; Uncharacterized protein | ||||
| STRING | Pavir.Ia00428.1.p | 0.0 | (Panicum virgatum) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G08150.1 | 2e-86 | KNOTTED-like from Arabidopsis thaliana | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pavir.9NG072500.3.p |



