 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pavir.9NG271400.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 1340aa MW: 148205 Da PI: 9.1911 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 11.2 | 0.0011 | 1304 | 1330 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH..T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt..H 23
y+Cp C ++F+ s++ rH r+ H
Pavir.9NG271400.1.p 1304 YVCPepGCEQTFRFVSDFSRHKRRtgH 1330
99********************98666 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00545 | 5.1E-14 | 18 | 59 | IPR003349 | JmjN domain |
| PROSITE profile | PS51183 | 13.536 | 19 | 60 | IPR003349 | JmjN domain |
| Pfam | PF02375 | 2.1E-12 | 20 | 53 | IPR003349 | JmjN domain |
| SMART | SM00558 | 3.4E-46 | 184 | 353 | IPR003347 | JmjC domain |
| PROSITE profile | PS51184 | 35.076 | 187 | 353 | IPR003347 | JmjC domain |
| SuperFamily | SSF51197 | 6.32E-25 | 200 | 351 | No hit | No description |
| Pfam | PF02373 | 1.3E-36 | 217 | 335 | IPR003347 | JmjC domain |
| Gene3D | G3DSA:3.30.160.60 | 4.5E-4 | 1214 | 1243 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 14 | 1221 | 1243 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 11.905 | 1244 | 1273 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 1.1 | 1244 | 1268 | IPR015880 | Zinc finger, C2H2-like |
| SuperFamily | SSF57667 | 4.63E-5 | 1246 | 1280 | No hit | No description |
| PROSITE pattern | PS00028 | 0 | 1246 | 1268 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 9.1E-4 | 1246 | 1272 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| Gene3D | G3DSA:3.30.160.60 | 1.7E-9 | 1273 | 1298 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 9.7E-4 | 1274 | 1298 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 11.718 | 1274 | 1303 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1276 | 1298 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 5.89E-10 | 1284 | 1328 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 2.2E-10 | 1299 | 1326 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.17 | 1304 | 1330 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.367 | 1304 | 1335 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1306 | 1330 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009826 | Biological Process | unidimensional cell growth | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0033169 | Biological Process | histone H3-K9 demethylation | ||||
| GO:0035067 | Biological Process | negative regulation of histone acetylation | ||||
| GO:0040010 | Biological Process | positive regulation of growth rate | ||||
| GO:0045815 | Biological Process | positive regulation of gene expression, epigenetic | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0071558 | Molecular Function | histone demethylase activity (H3-K27 specific) | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1340 aa Download sequence Send to blast |
MPSAQGELVP PWLRSLPLAP EFRPTAAEFA DPVAYLLKIE PAAAPFGICK VIPPLPPPPK 60 RTTLGNLSRS FAALHPGDPS PTFPTRHQEL GLCPRRPRPA LKPVWHSSRR YTLPQFEAAA 120 GVSRKALLAR LGVAASRQLS PLDVEALFWR STADRPVTVE YASDMPGSGF APCDARPTQL 180 PAAHVGETAW NMRGVARSPA SLLRFLREEV PGVTSPMLYV AMMFSWFAWH VEDHDLHSLN 240 YLHSGAPKTW YGVPRDAALA FEDAVRVHGY GGEVNPLETF AMLGDKTTVM SPEVLVRSGI 300 PCCRLVQNAG EFVVTFPGSY HSGFSHGFNY AEASNMATPE WLRVAKEAAV RRASINRPPM 360 VSHYQLLYEL ALSMCLRDPS SGAVEPRSSR LKEKKKGEGE QLVKKIFVQN VIEDNKLLNH 420 FLRDGSSCII LPTSPNDGSA LSTLLSKSQS TTKSRISDCH CNITEAPKDS RCLPVNGLLG 480 KNGELSSSKE LSTSVCSGEK FLPTACMHDC VNMSGSSDAN NAESDKGDIN STAGLLDQGF 540 LSCVTCGILS FSCVAVIKPR ECAAKWLMSA DSSLINKQFA DSGESHLIDA LQSATTNSGI 600 LRSGFEMDGN RTSSGAAALN RNSALDLLAS AYGDPSDSDE DVLNKKNQVA NVSSELISRT 660 IQSQPNNIST IGGCDGTNLS SSSKEHQQGP SSQRSQCIGN NNNGPKGVRT RNKYQLKMVL 720 SEGFQPKDKY SEIQKKVQCE PLSSNKASTE QLRGTDCQAG HNSATICMDS NTSTTTRVDN 780 LATSIVKPDK DSSRMHVFCL EHAIEVEKQL QTIGGAHIFL LCRPEYPKIE VEAKLLAEEV 840 EVEYDWKDIR FKETSIEDRK KMREVVQDEE TIPTNSDWAV KMGINLYYSA NLAKSPLYNK 900 QLPYNRVIYK AFGCSSPNNS PVKLKTYSRR QGRAKKIVLA GRWCGKVWMS NQVHPYLAHR 960 IKSHEPDEIN EICSSVQKSN SEHVEKSSRE ATCTRKSNSR AIEEKTSKRE EEPLEKANAN 1020 TPKHSEEDNS KALEGAAEAS AIKSSSRTVV EETSKRKKEP LEKANTKKPK HTEEENSKAL 1080 KGASEASHPS PSRMVIRSSS RIANRKNMLK SKMEEKDNDP ANRPKAMVEE DGNDPASCSR 1140 ARPLRQKTNT DVKKQTKKPR AEKQKAPSPT ALEDEDKELS FTKQQLSSRK QKTKVEEKQQ 1200 MKKTRENKGA PPSSPKQGEE YACNIEGCSM SFGTKQELSL HKRDICPVKG CGRKFFSHKY 1260 LLQHRKVHND DRPLKCSWKG CDMAFKWPWA RTEHMRVHTG DRPYVCPEPG CEQTFRFVSD 1320 FSRHKRRTGH AAKKAKTKK* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6ip0_A | 5e-71 | 12 | 374 | 8 | 353 | Transcription factor jumonji (Jmj) family protein |
| 6ip4_A | 5e-71 | 12 | 374 | 8 | 353 | Arabidopsis JMJ13 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 1054 | 1069 | KRKKEPLEKANTKKPK |
| 2 | 1324 | 1338 | KRRTGHAAKKAKTKK |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Pvr.26361 | 0.0 | callus | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in leaves and flag leaves. Expressed at low levels in roots, shoots, stems and panicles. {ECO:0000269|PubMed:24280387}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Histone demethylase that demethylates 'Lys-27' (H3K27me) of histone H3 with a specific activity for H3K27me3 and H3K27me2. No activity on H3K4me3, H3K9me3, H3K27me1 and H3K36me3. Involved in biotic stress response. May demethylate H3K27me3-marked defense-related genes and increase their basal and induced expression levels during pathogen infection. {ECO:0000269|PubMed:24280387}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00608 | ChIP-seq | Transfer from AT3G48430 | Download |
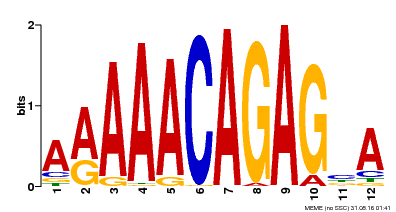
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pavir.9NG271400.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salt stress, abscisic acid (ABA), jasmonic acid (JA), the ethylene precursor ACC and infection by the bacterial pathogen Xanthomonas oryzae pv. oryzae. {ECO:0000269|PubMed:24280387}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025815151.1 | 0.0 | lysine-specific demethylase JMJ705-like | ||||
| Swissprot | Q5N712 | 0.0 | JM705_ORYSJ; Lysine-specific demethylase JMJ705 | ||||
| TrEMBL | A0A3L6SIW8 | 0.0 | A0A3L6SIW8_PANMI; Uncharacterized protein | ||||
| STRING | Pavir.Ea03512.1.p | 0.0 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP5346 | 32 | 46 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48430.1 | 0.0 | relative of early flowering 6 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pavir.9NG271400.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



