 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pavir.9NG288100.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 266aa MW: 28638 Da PI: 6.7915 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 61.3 | 2.2e-19 | 113 | 164 | 2 | 56 |
AP2 2 gykGVrwdkkrgrWvAeIrd.psengkrkrfslgkfgtaeeAakaaiaarkklege 56
y+GVr+++ +g+++AeIrd + k +r++lg+f taeeAa a+++a+++++g+
Pavir.9NG288100.1.p 113 SYRGVRRRP-WGKFAAEIRDtRR---KGARVWLGTFTTAEEAALAYDKAALRMRGP 164
69*******.**********332...34**************************95 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 1.33E-28 | 112 | 171 | No hit | No description |
| Pfam | PF00847 | 1.5E-14 | 113 | 163 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 1.2E-31 | 113 | 171 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 7.19E-23 | 113 | 173 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 2.7E-36 | 113 | 177 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 23.565 | 113 | 171 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 9.1E-11 | 114 | 125 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 9.1E-11 | 137 | 153 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 266 aa Download sequence Send to blast |
MAIGAPGSHQ PSNEAANILE NVWATIMTES ATPASSTAAS SEVGEEKPEA ILQRLPSLGR 60 WISMGAEEWD ELLLSGTALT SDASGELIVA SPASQDEDRG DRRAGSKAVA CKSYRGVRRR 120 PWGKFAAEIR DTRRKGARVW LGTFTTAEEA ALAYDKAALR MRGPRAHLNF PLDVVQRELA 180 GNSCVEASRV LRRRRRRGSN AAADTRSHGS VSATGGCDQT MVSFAFGKKD QGTSMVQERS 240 MSDPGAVIEF EDIGGEYWDY LFAPL* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2gcc_A | 7e-28 | 112 | 174 | 4 | 66 | ATERF1 |
| 3gcc_A | 7e-28 | 112 | 174 | 4 | 66 | ATERF1 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 188 | 196 | RVLRRRRRR |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00441 | DAP | Transfer from AT4G18450 | Download |
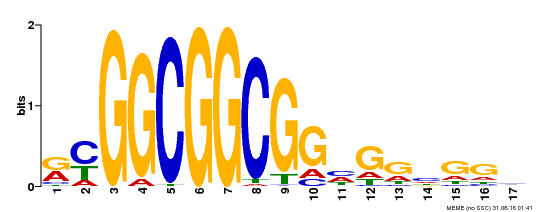
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pavir.9NG288100.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KU685412 | 1e-139 | KU685412.1 Saccharum spontaneum clone SES23E05, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025806647.1 | 1e-177 | ethylene-responsive transcription factor 15-like | ||||
| TrEMBL | A0A2S3H9P9 | 1e-176 | A0A2S3H9P9_9POAL; Uncharacterized protein | ||||
| STRING | Pavir.J34156.1.p | 0.0 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP11616 | 27 | 36 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G47220.1 | 3e-21 | ethylene responsive element binding factor 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pavir.9NG288100.1.p |



