 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pavir.9NG290100.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | RAV | ||||||||
| Protein Properties | Length: 386aa MW: 41159.7 Da PI: 10.2343 | ||||||||
| Description | RAV family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 39.5 | 1.4e-12 | 75 | 123 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
s+ykGV + +grW A+I++ r +r++lg+f + eAa+a++ a+++++g
Pavir.9NG290100.1.p 75 SKYKGVVPQP-NGRWGAQIYE------RhQRVWLGTFTGEAEAARAYDVAAQRFRG 123
89****9888.8*********......44**********99*************98 PP
| |||||||
| 2 | B3 | 94 | 1.1e-29 | 205 | 301 | 1 | 89 |
EEEE-..-HHHHTT-EE--HHH.HTT............---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT- CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh............ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegD 78
f+k+ tpsdv+k++rlv+pk++ae+h g + +++ l +ed g++W+++++y+++s++yvltkGW++Fvk++gL +gD
Pavir.9NG290100.1.p 205 FDKTVTPSDVGKLNRLVIPKQHAEKHfplqlpaaasvtG--ECKGVLLNFEDAAGKVWRFRYSYWNSSQSYVLTKGWSRFVKEKGLHAGD 292
89***************************8888766532..23889******************************************** PP
EEEEEE-SSSE CS
B3 79 fvvFkldgrse 89
v F+++ +
Pavir.9NG290100.1.p 293 AVGFYRSA--G 301
*****543..2 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 1.93E-23 | 75 | 131 | No hit | No description |
| Pfam | PF00847 | 2.6E-8 | 75 | 123 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 1.63E-16 | 75 | 131 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 5.9E-27 | 76 | 137 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 1.0E-19 | 76 | 131 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 19.862 | 76 | 131 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:2.40.330.10 | 3.3E-39 | 195 | 314 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 5.49E-29 | 203 | 303 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 3.06E-28 | 203 | 306 | No hit | No description |
| SMART | SM01019 | 1.0E-21 | 205 | 311 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 12.943 | 205 | 313 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 1.6E-26 | 205 | 301 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 386 aa Download sequence Send to blast |
MDSTSCLLDD LSSGGASTGK KQAPAPAAPS SRPLQRVGSG ASAVMDAAEP GAEADSGGAG 60 RAAAAGAVGG KLPSSKYKGV VPQPNGRWGA QIYERHQRVW LGTFTGEAEA ARAYDVAAQR 120 FRGRDAVTNF RPLSESDPES AAELRFLASR SKAEVVDMLR KHTYLEELAQ NRRAFAAASP 180 SPPPKNPASS PPSAASAAAA REHLFDKTVT PSDVGKLNRL VIPKQHAEKH FPLQLPAAAS 240 VTGECKGVLL NFEDAAGKVW RFRYSYWNSS QSYVLTKGWS RFVKEKGLHA GDAVGFYRSA 300 GKQLFIDCKV RPKTTSFANA ATTPPHAVKA VRLFGVDLLT PPKPTVAPEQ EEMLPMMTNK 360 RARDAIAAST PHMIFKKQCI DFALT* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wid_A | 4e-48 | 202 | 311 | 11 | 117 | DNA-binding protein RAV1 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Pvr.194 | 0.0 | root | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00025 | PBM | Transfer from AT1G68840 | Download |
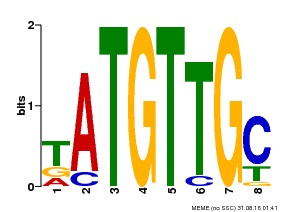
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pavir.9NG290100.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT041962 | 0.0 | BT041962.2 Zea mays full-length cDNA clone ZM_BFb0089O18 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025807648.1 | 0.0 | AP2/ERF and B3 domain-containing protein Os05g0549800 | ||||
| Swissprot | Q6L4H4 | 0.0 | Y5498_ORYSJ; AP2/ERF and B3 domain-containing protein Os05g0549800 | ||||
| TrEMBL | A0A2T7EAR2 | 0.0 | A0A2T7EAR2_9POAL; Uncharacterized protein | ||||
| STRING | Pavir.J13012.1.p | 0.0 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP456 | 38 | 203 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G68840.2 | 1e-102 | related to ABI3/VP1 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pavir.9NG290100.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



