 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pavir.J149500.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 642aa MW: 67117.2 Da PI: 7.3491 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 50.2 | 6.5e-16 | 300 | 359 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrd.pse.ng..kr.krfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV++++++gr++A+++d ++ g ++ ++++lg ++ +e+Aa+a++ a++k++g
Pavir.J149500.1.p 300 SQYRGVTRHRWTGRYEAHLWDnSCKkEGqtRKgRQVYLGGYDMEEKAARAYDLAALKYWG 359
78*******************988886678446*************************98 PP
| |||||||
| 2 | AP2 | 48.3 | 2.5e-15 | 402 | 453 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
s y+GV+++++ grW A+I +s +k +lg+f t eeAa+a++ a+ k++g
Pavir.J149500.1.p 402 SIYRGVTRHHQHGRWQARIGRVSG---NKDLYLGTFSTQEEAAEAYDVAAIKFRG 453
57****************999754...5************************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 1.9E-12 | 300 | 359 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 1.7E-16 | 300 | 368 | IPR016177 | DNA-binding domain |
| CDD | cd00018 | 7.98E-20 | 300 | 368 | No hit | No description |
| SMART | SM00380 | 8.7E-29 | 301 | 373 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 19.137 | 301 | 367 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 1.0E-15 | 301 | 367 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.5E-6 | 302 | 313 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 5.36E-18 | 402 | 463 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 1.9E-10 | 402 | 453 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 5.03E-24 | 402 | 463 | No hit | No description |
| SMART | SM00380 | 2.0E-35 | 403 | 467 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 19.098 | 403 | 461 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 4.0E-19 | 403 | 461 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.5E-6 | 443 | 463 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007276 | Biological Process | gamete generation | ||||
| GO:0010492 | Biological Process | maintenance of shoot apical meristem identity | ||||
| GO:0042127 | Biological Process | regulation of cell proliferation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 642 aa Download sequence Send to blast |
MTSNSSQNLS SCSTGGSDAA VGGGSWLGFS LSPHMAATMD GSNAVQVQQH HGGLFYPPVV 60 SSSPAGFYYA LGGGQDGVAT AGGNGGGGFY PGISSMPLKS DGSLCIMEAL HRSEQEHQGV 120 VVSSPSPKLE DFLGAGPAMA LSLDNSSYYY AGGRGHGHGQ DQGAYLQPLH CAVIPGSGAH 180 GHDVYGGHAQ LVDEQSAAAM AASWFSARGG GGYDVNGSGA ILPAQGLPHP LALSMSSGTG 240 SQSSCVTMQV GAHPDPHADA VAEYIAMDGN GSKKRGGAGA LQKQPTVHRK SIDTFGQRTS 300 QYRGVTRHRW TGRYEAHLWD NSCKKEGQTR KGRQVYLGGY DMEEKAARAY DLAALKYWGT 360 STHINFPIED YREELEEMKN MTRQEYVAHL RRKSSGFSRG ASIYRGVTRH HQHGRWQARI 420 GRVSGNKDLY LGTFSTQEEA AEAYDVAAIK FRGQSAVTNF DISRYDVDKI LESNTLLPGE 480 QVRRRKGGEA GGAVAESDAV ARATAALEQA GNGAAADTWR IQAARADGLH GQQQHQGLLS 540 SEAFSLLHDI VSVDTAGHGG GGASAHMSNA SSLAPSVSNS REQSPDRGGG GLAMLFSKPS 600 AAASKLACPL PLGSWASPSP VSARPGVSIA HLPMFAAWTD A* |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Expressed in floral primordia, in STM-negative region, then in sepal primordia. As sepal develops, progressively confined to a basal core before disappearing. Present in stamen primordia, then confined to a central region as they become stalked and develop locules. Later reduced to procambial cells as stamen mature. From petal primordia, expressed on the lateral edges of developing petals and finally confined to petal epidermis before disappearing. Present in carpel primordia, then in inner side of carpels especially in the placenta. Strong levels in ovules primordia and young ovules, then localized in integuments initiation zone before being confined to inner integument cells that will differentiate into the endothelium. Expressed in the distal half of the funiculus throughout ovule development and later extends into the chalaza. After fertilization, expression shift to the embryo. First on the apical part at the globular stage, then in cotyledons primordia, and later in cotyledons during the torpedo stage. As cotyledons grow out, expression becomes limited to a plane separating adaxial and abaxial parts. Excluded from the embryonic central region (ECR). In seedlings, found in leaf primordia then in central and lateral actively developing regions of extending leaves. {ECO:0000269|PubMed:10656774, ECO:0000269|PubMed:8742707, ECO:0000269|PubMed:9671577}. | |||||
| Uniprot | TISSUE SPECIFICITY: Mostly expressed in developing flowers. Also present in mature flowers, siliques and seedlings, but not in mature roots, leaves and stems. Expressed in ovules and in vegetative and floral primordia. {ECO:0000269|PubMed:15988559, ECO:0000269|PubMed:8742706, ECO:0000269|PubMed:8742707}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that recognizes and binds to the DNA consensus sequence 5'-CAC[AG]N[AT]TNCCNANG-3'. Required for the initiation and growth of ovules integumenta, and for the development of female gametophyte. Plays a critical role in the development of gynoecium marginal tissues (e.g. stigma, style and septa), and in the fusion of carpels and of medial ridges leading to ovule primordia. Also involved in organs initiation and development, including floral organs. Maintains the meristematic competence of cells and consequently sustains expression of cell cycle regulators during organogenesis, thus controlling the final size of each organ by controlling their cell number. Regulates INO autoinduction and expression pattern. As ANT promotes petal cell identity and mediates down-regulation of AG in flower whorl 2, it functions as a class A homeotic gene. {ECO:0000269|PubMed:10528263, ECO:0000269|PubMed:10639184, ECO:0000269|PubMed:10948255, ECO:0000269|PubMed:11041883, ECO:0000269|PubMed:12183381, ECO:0000269|PubMed:12271029, ECO:0000269|PubMed:12655002, ECO:0000269|PubMed:8742706, ECO:0000269|PubMed:8742707, ECO:0000269|PubMed:9001406, ECO:0000269|PubMed:9093862, ECO:0000269|PubMed:9118807}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00088 | SELEX | Transfer from AT4G37750 | Download |
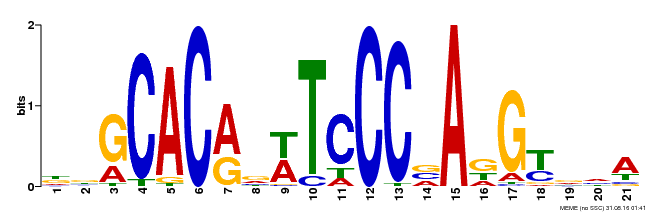
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pavir.J149500.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK375318 | 0.0 | AK375318.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv3090N20. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025796942.1 | 0.0 | AP2-like ethylene-responsive transcription factor CRL5 | ||||
| Swissprot | Q38914 | 1e-142 | ANT_ARATH; AP2-like ethylene-responsive transcription factor ANT | ||||
| TrEMBL | A0A1E5VRG8 | 0.0 | A0A1E5VRG8_9POAL; AP2-like ethylene-responsive transcription factor ANT | ||||
| STRING | Pavir.J24038.1.p | 0.0 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP948 | 38 | 133 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G37750.1 | 1e-126 | AP2 family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pavir.J149500.1.p |



