 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pbr010932.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Maleae; Pyrus
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 374aa MW: 41605 Da PI: 7.7921 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 100.2 | 1.3e-31 | 76 | 129 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
rlrWtp+LH +Fv+ave+LGG+e+AtPk +l+lm+vkgL+++hvkSHLQ+YR+
Pbr010932.1 76 SRLRWTPDLHLCFVRAVERLGGQERATPKLVLQLMNVKGLSIAHVKSHLQMYRS 129
69***************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 12.565 | 72 | 132 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 8.1E-30 | 72 | 130 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 5.55E-15 | 73 | 131 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 5.4E-23 | 76 | 131 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 3.3E-9 | 77 | 128 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 374 aa Download sequence Send to blast |
MKSESTSIEE VSKSSPSTSK KEYDHEDDEG EEDEVDEDGL QLKNNGDSSS NSTIEENHEK 60 KGASGSVRHY VRSKTSRLRW TPDLHLCFVR AVERLGGQER ATPKLVLQLM NVKGLSIAHV 120 KSHLQMYRSK KTEDPNQVLT DQAGFFMEGG DNQLYNLTQL SMLQSFNRWS SSGLRYGAND 180 HSSWRGHHHQ IYSPYSSSRT SALLDHNTRN IGLYGSVAER ILGSNSNNIM TTSANQLYTN 240 LPSPTHGQTT WRRNRDELCQ TMIHRSCQDH HSWTAAIRDH QGQNMLKRKG LDTDNCDLDL 300 NLSLKVPPKH DHGFGKGLQY GGDNHQKLLM GCSLSLTLSS SSSSKLGVKK LEGTGHGDGK 360 HGKNMASTLD LTL* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4r_A | 7e-18 | 76 | 131 | 2 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 7e-18 | 76 | 131 | 2 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 7e-18 | 76 | 131 | 2 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 7e-18 | 76 | 131 | 2 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00301 | DAP | Transfer from AT2G40260 | Download |
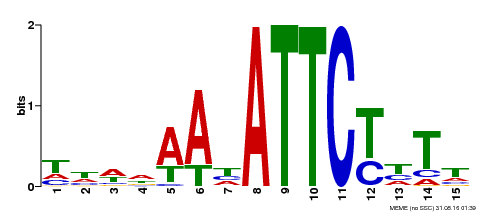
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pbr010932.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009369202.1 | 0.0 | PREDICTED: uncharacterized protein LOC103958635 | ||||
| TrEMBL | A0A498JM49 | 0.0 | A0A498JM49_MALDO; Uncharacterized protein | ||||
| STRING | XP_009369202.1 | 0.0 | (Pyrus x bretschneideri) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2378 | 34 | 82 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G38300.1 | 1e-46 | G2-like family protein | ||||



