 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pbr012024.1 | ||||||||
| Common Name | ERF2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Maleae; Pyrus
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 272aa MW: 29815.5 Da PI: 7.5587 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 65.2 | 1.3e-20 | 89 | 138 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
y+G+r+++ +g+W+AeIrdp + r++lg+++taeeAa+a+++a+ +++g
Pbr012024.1 89 VYRGIRQRP-WGKWAAEIRDPH----KgVRVWLGTYDTAEEAARAYDEAAVRIRG 138
69*******.**********72....35**********************99997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00380 | 3.9E-39 | 89 | 152 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 24.131 | 89 | 146 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 5.6E-14 | 89 | 138 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 1.31E-22 | 89 | 146 | IPR016177 | DNA-binding domain |
| Gene3D | G3DSA:3.30.730.10 | 8.8E-33 | 89 | 146 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 6.8E-12 | 90 | 101 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 2.02E-32 | 90 | 146 | No hit | No description |
| PRINTS | PR00367 | 6.8E-12 | 112 | 128 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0008219 | Biological Process | cell death | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009735 | Biological Process | response to cytokinin | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0010286 | Biological Process | heat acclimation | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0051707 | Biological Process | response to other organism | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005737 | Cellular Component | cytoplasm | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 272 aa Download sequence Send to blast |
MCGGAIISDF IAAKRGRKLT AQDLWSDLDT ISDLLGIDHS NSINKQPENH KVVQKPKPSI 60 TKVVTSDEKP KKASGSAAAA EGKRVRKNVY RGIRQRPWGK WAAEIRDPHK GVRVWLGTYD 120 TAEEAARAYD EAAVRIRGDK AKLNFAQPPP SSPLPSLAPD TPPPTKRRCI VAESTRVEPT 180 QPSFQTGSYY YDPLYHGGGG GEMYAKNEVA GGDGRYELKE QIWSLESFLG LDEAVVEQPS 240 QVVSGSGESD SLDLWMLDDL VAYRQQGQLL Y* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2gcc_A | 9e-22 | 90 | 145 | 6 | 62 | ATERF1 |
| 3gcc_A | 9e-22 | 90 | 145 | 6 | 62 | ATERF1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00109 | PBM | Transfer from AT3G16770 | Download |
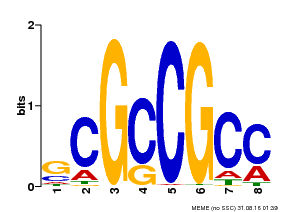
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pbr012024.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KJ623716 | 0.0 | KJ623716.1 Pyrus x bretschneideri ethylene response factor 2 (ERF2) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009370001.1 | 0.0 | PREDICTED: ethylene-responsive transcription factor RAP2-3 isoform X1 | ||||
| TrEMBL | A0A0D3QIW3 | 0.0 | A0A0D3QIW3_9ROSA; Ethylene response factor 2 | ||||
| STRING | XP_009370001.1 | 0.0 | (Pyrus x bretschneideri) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4892 | 33 | 53 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G16770.1 | 3e-43 | ethylene-responsive element binding protein | ||||



