 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pbr013255.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Maleae; Pyrus
|
||||||||
| Family | RAV | ||||||||
| Protein Properties | Length: 387aa MW: 42451.5 Da PI: 9.2363 | ||||||||
| Description | RAV family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 43.2 | 9.5e-14 | 76 | 124 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
s+ykGV + +grW A+I++ ++r++lg+f ++eAa+a++ a+++++g
Pbr013255.1 76 SKYKGVVPQP-NGRWGAQIYEK-----HQRVWLGTFNEEDEAARAYDVAAQRFRG 124
89****9888.8*********3.....5**********99*************98 PP
| |||||||
| 2 | B3 | 96.5 | 1.7e-30 | 213 | 322 | 1 | 96 |
EEEE-..-HHHHTT-EE--HHH.HTT..............---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh..............ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkl 84
f+k+ tpsdv+k++rlv+pk++ae+h ++ +++ l +ed g++W+++++y+++s++yvltkGW++Fvk+++Lk+gD+v F++
Pbr013255.1 213 FEKAVTPSDVGKLNRLVIPKQHAEKHfplqsgstatvtvsASSACKGVLLNFEDVGGKVWRFRYSYWNSSQSYVLTKGWSRFVKEKNLKAGDIVSFQR 310
89*************************999766666655533344999*************************************************8 PP
-SSSEE..EEEE CS
B3 85 dgrsefelvvkv 96
+ +++l++ +
Pbr013255.1 311 STGPDKQLYIDW 322
766777777765 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF54171 | 9.81E-17 | 76 | 132 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 3.0E-9 | 76 | 124 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 2.96E-25 | 76 | 134 | No hit | No description |
| SMART | SM00380 | 9.0E-29 | 77 | 138 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 19.862 | 77 | 132 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 1.1E-19 | 77 | 132 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:2.40.330.10 | 2.5E-40 | 207 | 329 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 1.11E-29 | 211 | 325 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 1.53E-28 | 212 | 311 | No hit | No description |
| Pfam | PF02362 | 8.2E-28 | 213 | 322 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 1.2E-23 | 213 | 326 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 13.084 | 213 | 325 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0048573 | Biological Process | photoperiodism, flowering | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 387 aa Download sequence Send to blast |
MDGISSTEES TTSDSISISP PHHIVTRVEP LAKSAPQVDS LCRVGSGASS VILDSDLSSS 60 GGTGGVEAES RKLPSSKYKG VVPQPNGRWG AQIYEKHQRV WLGTFNEEDE AARAYDVAAQ 120 RFRGRDAVTN LKPSSAEPIF SDDEENDDVE AVFLSSHSKS EIVDMLRKHT YNDELEQSKR 180 NYCAYGKRGR SNGPLGLFGM DNSRVQKARE QLFEKAVTPS DVGKLNRLVI PKQHAEKHFP 240 LQSGSTATVT VSASSACKGV LLNFEDVGGK VWRFRYSYWN SSQSYVLTKG WSRFVKEKNL 300 KAGDIVSFQR STGPDKQLYI DWKTRMSVNN NSNGSNPVQG QVAPVQMVRL FGVNIFKIPG 360 SVGAGPVDAA AAAXXXXCRC YWRRLQ* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wid_A | 2e-57 | 210 | 334 | 11 | 126 | DNA-binding protein RAV1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional repressor of flowering time on long day plants. Acts directly on FT expression by binding 5'-CAACA-3' and 5'-CACCTG-3 sequences. Functionally redundant with TEM2. {ECO:0000269|PubMed:18718758}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00024 | PBM | Transfer from AT1G25560 | Download |
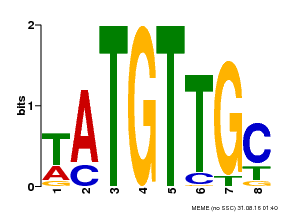
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pbr013255.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Expressed with a circadian rhythm showing a peak at dawn. {ECO:0000269|PubMed:18718758}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | GU732460 | 0.0 | GU732460.1 Malus x domestica AP2 domain class transcription factor (AP2D36) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009356262.1 | 0.0 | PREDICTED: LOW QUALITY PROTEIN: AP2/ERF and B3 domain-containing transcription factor RAV1-like | ||||
| Swissprot | Q9C6M5 | 1e-137 | RAVL1_ARATH; AP2/ERF and B3 domain-containing transcription repressor TEM1 | ||||
| TrEMBL | A0A482LZ61 | 0.0 | A0A482LZ61_9ROSA; Transcription factor RAV1 | ||||
| STRING | XP_009356262.1 | 0.0 | (Pyrus x bretschneideri) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2920 | 32 | 74 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G13260.1 | 1e-126 | related to ABI3/VP1 1 | ||||



