 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pbr014125.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Maleae; Pyrus
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 356aa MW: 39472.5 Da PI: 4.834 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 59.4 | 8.8e-19 | 122 | 172 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
++++GVr+++ +g+W+AeIrdp r r +lg+f+taeeAa +++a+ +l+g
Pbr014125.1 122 KKFRGVRQRP-WGKWAAEIRDPL----RrVRLWLGTFDTAEEAAVVYDNAAIQLRG 172
59********.**********72....45*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 8.10E-32 | 122 | 180 | No hit | No description |
| SMART | SM00380 | 1.1E-37 | 123 | 186 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 23.578 | 123 | 180 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 4.0E-11 | 123 | 172 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 2.55E-21 | 123 | 180 | IPR016177 | DNA-binding domain |
| Gene3D | G3DSA:3.30.730.10 | 4.5E-30 | 123 | 180 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.0E-10 | 124 | 135 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.0E-10 | 146 | 162 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0042991 | Biological Process | transcription factor import into nucleus | ||||
| GO:0048364 | Biological Process | root development | ||||
| GO:0048825 | Biological Process | cotyledon development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 356 aa Download sequence Send to blast |
MNRPAVRYTH HRSHTTMVAN SAEADDFESV RPRVVRISVT DGNATDSSSD DEAEGSSFCG 60 TRHRVKRFVN EITIESCSNR DTDSAVWRSR MSRTGRKRPS GKSKGPASGR RDLKAVAAPA 120 GKKFRGVRQR PWGKWAAEIR DPLRRVRLWL GTFDTAEEAA VVYDNAAIQL RGPDALTNFA 180 PPPPAISLPD VKPEVSSGFY SGDGSHNHLC SPTSVLRCPS PSNEEAESPW PKQSPEVTEV 240 RNVRENSSIS KSLSEFSDYT SFETFIPNDI FNFETSIPDF FEENNLRDDI CISKEEITDF 300 GNFGISKEEM TDFGNLGDLF VDSVQDFGFG SSPWCTEDYF QDIGDLFSSD PLVAL* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2gcc_A | 7e-20 | 121 | 179 | 3 | 62 | ATERF1 |
| 3gcc_A | 7e-20 | 121 | 179 | 3 | 62 | ATERF1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00027 | PBM | Transfer from AT4G23750 | Download |
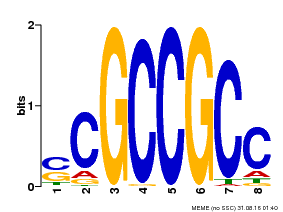
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pbr014125.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009352357.1 | 0.0 | PREDICTED: ethylene-responsive transcription factor CRF2 | ||||
| TrEMBL | A0A498JPC1 | 0.0 | A0A498JPC1_MALDO; Uncharacterized protein | ||||
| STRING | XP_009352357.1 | 0.0 | (Pyrus x bretschneideri) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF450 | 34 | 161 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G23750.2 | 9e-64 | cytokinin response factor 2 | ||||



