 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pbr015587.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Maleae; Pyrus
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 320aa MW: 34900.2 Da PI: 10.263 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 40.9 | 4.7e-13 | 130 | 174 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT+eE+ ++ + ++lG+g+W+ I+r ++Rt+ q+ s+ qky
Pbr015587.1 130 PWTEEEHRTFLIGLEKLGKGDWRGISRNYVTTRTPTQVASHAQKY 174
8*******************************************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50158 | 8.598 | 3 | 20 | IPR001878 | Zinc finger, CCHC-type |
| PROSITE profile | PS51294 | 19.284 | 123 | 179 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.2E-17 | 124 | 180 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 3.8E-18 | 126 | 178 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 2.8E-9 | 127 | 177 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 7.0E-11 | 127 | 173 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.46E-8 | 130 | 175 | No hit | No description |
| Pfam | PF00249 | 1.1E-10 | 130 | 174 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 320 aa Download sequence Send to blast |
MGRKCSHCGN VGHNSRTCTN FRGTLSSTSF VGFRLFGVQL QHDHHHVVMS SSNMNSNSAN 60 HHVAMSMKKS FSTDCLPTTS SASSSSPSNS SLSPSRLSMD ENICAADKAS IGYLSDGLIC 120 RAQERKKGVP WTEEEHRTFL IGLEKLGKGD WRGISRNYVT TRTPTQVASH AQKYFLRQAS 180 LTKKKRRSSL FDMFGSSNNI NMATYDPIPT QRDNIIHGDM AIDTSTLPPL QRSTAAKNLK 240 SDHNQKPQSS RTTTQQLPAW IYGLIDSQMK SSSSSSEAPV VPDLELTLAA PTSPNLEQKK 300 RSSPTPAGPL LNLGRISVT* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 182 | 186 | KKKRR |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00565 | DAP | Transfer from AT5G56840 | Download |
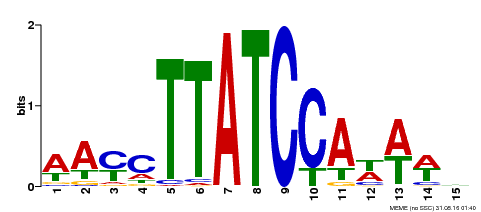
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pbr015587.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009352631.1 | 0.0 | PREDICTED: myb-like protein J | ||||
| TrEMBL | A0A498HTD9 | 0.0 | A0A498HTD9_MALDO; Uncharacterized protein | ||||
| STRING | XP_009352631.1 | 0.0 | (Pyrus x bretschneideri) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3213 | 32 | 71 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G56840.1 | 9e-57 | MYB_related family protein | ||||



