 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pbr016089.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Maleae; Pyrus
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 332aa MW: 35284.3 Da PI: 7.9779 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 211 | 3.1e-65 | 3 | 153 | 1 | 149 |
DUF822 1 ggsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeaeaagssasaspesslqssl 98
++++r+ptwkErEnnk+RERr a aakiy+GLR++Gnyklpk++DnneVlkALc+eAGw+ve+DGttyrkg+kp ++++++g s+sasp+ss+q s+
Pbr016089.1 3 TSGTRMPTWKERENNKKRERRGPATAAKIYSGLRMYGNYKLPKHCDNNEVLKALCNEAGWTVEEDGTTYRKGCKPVDRMDIMGGSTSASPCSSYQPSP 100
5899********************************************************************************************** PP
DUF822 99 kssalaspvesysaspksssfpspssldsislasa..asllpvlsvlslvsss 149
+s+ +sp +sy++sp+sssfpsp+++ +++ +a +sl+p+l++ls+ sss
Pbr016089.1 101 CASYNPSPGASYNPSPASSSFPSPARYHANENGNAdaNSLIPWLKTLSSGSSS 153
**************************99888766566**********997766 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 1.1E-64 | 4 | 148 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 332 aa Download sequence Send to blast |
MMTSGTRMPT WKERENNKKR ERRGPATAAK IYSGLRMYGN YKLPKHCDNN EVLKALCNEA 60 GWTVEEDGTT YRKGCKPVDR MDIMGGSTSA SPCSSYQPSP CASYNPSPGA SYNPSPASSS 120 FPSPARYHAN ENGNADANSL IPWLKTLSSG SSSAMSKLPQ FFVQGGSISA PVTPPLSSPT 180 CRTPRTKSDW DEAVAGSSWA GQHYPFLPSS TPPSPGRQVL PDSGWLAGLQ IPQSGPSSPT 240 FSLVSKNPFN LKEALSGAGS RMWTPGQSGT CSPVVAASID HTGDIPMSDG MAAEFAFGSN 300 TTGLVKPWEG ERIEECVSDD LELTLGSTRT R* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5zd4_A | 5e-24 | 7 | 78 | 372 | 443 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_B | 5e-24 | 7 | 78 | 372 | 443 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_C | 5e-24 | 7 | 78 | 372 | 443 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_D | 5e-24 | 7 | 78 | 372 | 443 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00248 | DAP | Transfer from AT1G78700 | Download |
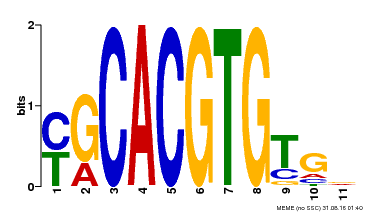
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pbr016089.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009352718.1 | 0.0 | PREDICTED: BES1/BZR1 homolog protein 4-like | ||||
| Swissprot | Q9ZV88 | 1e-126 | BEH4_ARATH; BES1/BZR1 homolog protein 4 | ||||
| TrEMBL | A0A498IT00 | 0.0 | A0A498IT00_MALDO; Uncharacterized protein | ||||
| STRING | XP_009352718.1 | 0.0 | (Pyrus x bretschneideri) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2661 | 33 | 75 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G78700.1 | 1e-115 | BES1/BZR1 homolog 4 | ||||



