 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pbr018624.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Maleae; Pyrus
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 280aa MW: 30528 Da PI: 4.8212 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 60.8 | 3.1e-19 | 40 | 89 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
++ GVr+++ +gr++AeIrdps+ + r++lg+f+taeeAa a+++a+++++g
Pbr018624.1 40 RFLGVRRRP-WGRYAAEIRDPST---KERHWLGTFDTAEEAALAYDRAARSMRG 89
678******.**********965...5*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 7.35E-31 | 39 | 96 | No hit | No description |
| PROSITE profile | PS51032 | 20.705 | 40 | 97 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 6.6E-35 | 40 | 103 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 2.9E-29 | 41 | 97 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 4.9E-21 | 41 | 98 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 4.1E-12 | 41 | 89 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.5E-8 | 41 | 52 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.5E-8 | 63 | 79 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 280 aa Download sequence Send to blast |
MDFFTASPSK SNNSPSSSSK TKRKQQQQQT QQPQTQQETR FLGVRRRPWG RYAAEIRDPS 60 TKERHWLGTF DTAEEAALAY DRAARSMRGS KARTNFVYSD MPHGSSVTSI ISPDESQRDM 120 SAFFTNPLPE THSQSQTHTQ QPFFTHQDPF DAYTVSGGSS TEFGSGSYGP TPGVVADESG 180 YQSESHTHEL PPLPPCVSSS TCYGPEGIMS SDGVWNESGF YGFSEPTTNN GLDSMVSGSY 240 VGFDSNEFVQ HSPLFGSMPP VSDAGIDGFD LGSSSAYFY* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gcc_A | 7e-20 | 41 | 96 | 3 | 59 | ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). {ECO:0000250}. | |||||
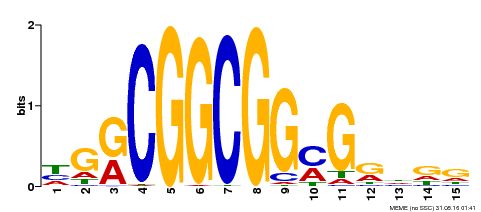
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00163 | DAP | Transfer from AT1G28160 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pbr018624.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009353495.1 | 0.0 | PREDICTED: ethylene-responsive transcription factor LEP-like | ||||
| Swissprot | Q9FZ90 | 2e-51 | ERF87_ARATH; Ethylene-responsive transcription factor ERF087 | ||||
| TrEMBL | A0A498HP73 | 1e-145 | A0A498HP73_MALDO; Uncharacterized protein | ||||
| STRING | XP_009353495.1 | 0.0 | (Pyrus x bretschneideri) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3904 | 31 | 61 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G28160.1 | 1e-38 | ERF family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



