 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pbr022508.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Maleae; Pyrus
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 545aa MW: 59648.9 Da PI: 6.8964 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 88.4 | 6.6e-28 | 228 | 282 | 1 | 56 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRla 56
k +++WtpeLH+rFv+aveqL G +kA+P++ilelm+v++Lt+++++SHLQkYR++
Pbr022508.1 228 KVKVDWTPELHRRFVQAVEQL-GVDKAVPSRILELMGVDCLTRHNIASHLQKYRSH 282
5689*****************.********************************86 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 18.908 | 225 | 284 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 4.2E-27 | 226 | 285 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 2.51E-18 | 226 | 285 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.1E-26 | 228 | 282 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 6.3E-8 | 231 | 280 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0007165 | Biological Process | signal transduction | ||||
| GO:0009658 | Biological Process | chloroplast organization | ||||
| GO:0009910 | Biological Process | negative regulation of flower development | ||||
| GO:0010380 | Biological Process | regulation of chlorophyll biosynthetic process | ||||
| GO:0010638 | Biological Process | positive regulation of organelle organization | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:1900056 | Biological Process | negative regulation of leaf senescence | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 545 aa Download sequence Send to blast |
MLLLSPLTEG RHHHQLKDEK QLGDDGDVES SFAFNGGNTT VLDFPEFSGG MGMGNLLDSI 60 DFDDLFIGIH DGDVLPDLEM DSEILDFSVP APADDINTKT TPPPPSKEEE EVADDYYNTT 120 KTTTTSSKVE DQEEEDLHGH GEVAAAAVSI TSTNNNSGLN QNYSSSSTTT PPDNRGDQEI 180 VSKRDDINQI RAAYRPSSGS TRRKSADHKL ATKSSPTAQS KKSHGKRKVK VDWTPELHRR 240 FVQAVEQLGV DKAVPSRILE LMGVDCLTRH NIASHLQKYR SHRKHLLARE AEAASWTQRR 300 QMYGAAAAAG GGAGPKSRRD VMMMNNPNWL NAPTIGFPSI TSTTPPPMHH AQHHRQLIRP 360 LHVWGHPTMD QSMMHMWPPK HHLAHHFPPP VLPSPPPAPA HAWPPGPPPP PDASYWHHPH 420 SHHQRAPDAP TPGAPCFPRQ QLAAPTVTNH SLQQRFHTPP VPGIPPHAMY KVEPAGIAAP 480 TPPQSGLHLL LDFHPSKESI DAAIGDVLSK PWLPLPLGLR PPATDTVMVE LQRQGVQKFP 540 PSCA* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 2e-15 | 225 | 280 | 2 | 57 | ARR10-B |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00022 | PBM | Transfer from AT2G20570 | Download |
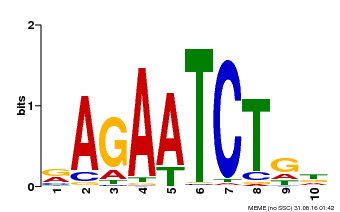
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pbr022508.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018502530.1 | 0.0 | PREDICTED: probable transcription factor GLK1 isoform X1 | ||||
| TrEMBL | A0A498HB96 | 0.0 | A0A498HB96_MALDO; Uncharacterized protein | ||||
| STRING | XP_009354290.1 | 0.0 | (Pyrus x bretschneideri) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4242 | 34 | 61 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G20570.1 | 4e-76 | GBF's pro-rich region-interacting factor 1 | ||||



