 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pbr025856.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Maleae; Pyrus
|
||||||||
| Family | TCP | ||||||||
| Protein Properties | Length: 441aa MW: 47799 Da PI: 7.0474 | ||||||||
| Description | TCP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | TCP | 105.3 | 9.9e-33 | 47 | 161 | 1 | 89 |
TCP 1 aagkkdrhskihTkvggRdRRvRlsaecaarfFdLqdeLGfdkdsktieWLlqqakpaikeltgt.......................ssssasecea 75
a+g+kdrhsk++T++g+RdRRvRl a++a++f+d+qd+LG+d++sk+++WL+++ak+ai+el ++
Pbr025856.1 47 ATGRKDRHSKVCTAKGPRDRRVRLAAHTAIQFYDVQDRLGYDRPSKAVDWLIKKAKAAIDELEELpswnphsisttaavpametqnpsA--------- 135
589**************************************************************665555444444444444333330......... PP
TCP 76 esssssasnsssg........................k 89
+ +
Pbr025856.1 136 ------------TgihcfaavdaigsanrrtmvgsgvS 161
............13334444444443333333322110 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03634 | 6.7E-29 | 48 | 224 | IPR005333 | Transcription factor, TCP |
| PROSITE profile | PS51369 | 33.273 | 50 | 108 | IPR017887 | Transcription factor TCP subgroup |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009793 | Biological Process | embryo development ending in seed dormancy | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0030154 | Biological Process | cell differentiation | ||||
| GO:0045962 | Biological Process | positive regulation of development, heterochronic | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 441 aa Download sequence Send to blast |
MEDSHHHHLN NHQATTSSRL GIRPPSSGGG VGTDIVEVVR GSHIVRATGR KDRHSKVCTA 60 KGPRDRRVRL AAHTAIQFYD VQDRLGYDRP SKAVDWLIKK AKAAIDELEE LPSWNPHSIS 120 TTAAVPAMET QNPSATGIHC FAAVDAIGSA NRRTMVGSGV SEQQFAQNPN SNSTFLPPSL 180 DSDAIADTIK SFFPMGASEA TAAAEAPSST IQFHQNYPPD LLSKASSQSQ DLRLSLQSFQ 240 DPILLHHHDS QNQHHHAQTH QNEQTLFSGS QNQLGFDGTS GSWAEHHHNQ QQQEMNRYQR 300 MVAWNSGGGD SGGNGGPSSA GGFIFNSLLP TQHGSTSPLQ QQQQSLFGQS QFFSQRGPLQ 360 SSNSPSVRAW MMDQQNQQSI SHDHHHQISQ SIHHQQSISG MGFASGGGFS GFHIPARIHG 420 EEEHDGTSDK PSSASSNSRH * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5zkt_A | 2e-20 | 55 | 108 | 1 | 54 | Putative transcription factor PCF6 |
| 5zkt_B | 2e-20 | 55 | 108 | 1 | 54 | Putative transcription factor PCF6 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor playing a pivotal role in the control of morphogenesis of shoot organs by negatively regulating the expression of boundary-specific genes such as CUC genes, probably through the induction of miRNA (e.g. miR164) (PubMed:12931144, PubMed:17307931). Required during early steps of embryogenesis (PubMed:15634699). Participates in ovule develpment (PubMed:25378179). Activates LOX2 expression by binding to the 5'-GGACCA-3' motif found in its promoter (PubMed:18816164). {ECO:0000269|PubMed:12931144, ECO:0000269|PubMed:15634699, ECO:0000269|PubMed:17307931, ECO:0000269|PubMed:18816164, ECO:0000269|PubMed:25378179}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00009 | PBM | Transfer from 929286 | Download |
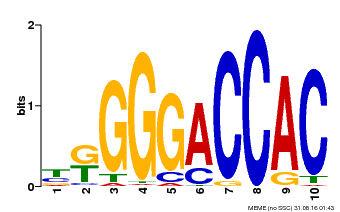
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pbr025856.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by the miRNA miR-JAW/miR319 (PubMed:12931144, PubMed:18816164). {ECO:0000269|PubMed:12931144, ECO:0000269|PubMed:18816164}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AB531022 | 0.0 | AB531022.1 Malus x domestica MdTCP4B mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009335205.1 | 0.0 | PREDICTED: transcription factor TCP4-like | ||||
| Swissprot | Q8LPR5 | 1e-108 | TCP4_ARATH; Transcription factor TCP4 | ||||
| TrEMBL | D9ZJD0 | 0.0 | D9ZJD0_MALDO; TCP domain class transcription factor | ||||
| STRING | XP_009335205.1 | 0.0 | (Pyrus x bretschneideri) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1163 | 34 | 105 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G15030.3 | 8e-75 | TCP family protein | ||||



