 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pbr030607.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Maleae; Pyrus
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 178aa MW: 19438.1 Da PI: 6.6647 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 48.9 | 1.6e-15 | 114 | 161 | 2 | 53 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkl 53
+y+GV++ + +g+WvAeIr+p++ ++ +lg+f tae+Aa a++ a+ l
Pbr030607.1 114 RYRGVHQQS-KGKWVAEIREPRK---CTCKWLGTFATAEDAACAYDLAAIIL 161
59****999.7*********854...8*********************9765 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 8.1E-8 | 114 | 158 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 9.91E-23 | 114 | 173 | No hit | No description |
| PROSITE profile | PS51032 | 17.886 | 114 | 171 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 8.5E-17 | 114 | 171 | IPR016177 | DNA-binding domain |
| Gene3D | G3DSA:3.30.730.10 | 7.4E-23 | 114 | 171 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 1.7E-25 | 114 | 177 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.4E-5 | 115 | 126 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.4E-5 | 137 | 153 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006970 | Biological Process | response to osmotic stress | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009738 | Biological Process | abscisic acid-activated signaling pathway | ||||
| GO:0009744 | Biological Process | response to sucrose | ||||
| GO:0009749 | Biological Process | response to glucose | ||||
| GO:0010119 | Biological Process | regulation of stomatal movement | ||||
| GO:0010353 | Biological Process | response to trehalose | ||||
| GO:0010449 | Biological Process | root meristem growth | ||||
| GO:0010896 | Biological Process | regulation of triglyceride catabolic process | ||||
| GO:0031930 | Biological Process | mitochondria-nucleus signaling pathway | ||||
| GO:0032880 | Biological Process | regulation of protein localization | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0048316 | Biological Process | seed development | ||||
| GO:0048527 | Biological Process | lateral root development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 178 aa Download sequence Send to blast |
MSTSEDEEVE ELEDDDIIGK ALQKCAKISV DLRKKLQGSS ALAVSDWYTE VEAASVKIVN 60 QAITYLMLLK HLNDDLGPHL IVCPASVLEI GKENLKSSAH HSFSKGGPDN KKFRYRGVHQ 120 QSKGKWVAEI REPRKCTCKW LGTFATAEDA ACAYDLAAII LYASRAQLNL QPSGSSS* |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00674 | SELEX | Transfer from GRMZM2G093595 | Download |
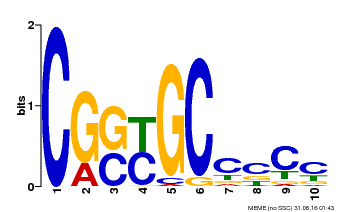
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pbr030607.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF8847 | 29 | 40 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G40220.1 | 7e-21 | ERF family protein | ||||



