 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pbr035774.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Maleae; Pyrus
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 302aa MW: 33867.7 Da PI: 6.6932 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 65.3 | 1.2e-20 | 191 | 242 | 2 | 56 |
AP2 2 gykGVrwdkkrgrWvAeIrdpseng.krkrfslgkfgtaeeAakaaiaarkklege 56
+y+GVr+++ +g+++AeIrdp + + r++lg+f+ta eAa+a+++a+ kl+g+
Pbr035774.1 191 HYRGVRQRP-WGKYAAEIRDP---NrRGSRVWLGTFDTAIEAARAYDRAAFKLRGA 242
8********.**********9...4334**************************95 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 2.08E-30 | 190 | 249 | No hit | No description |
| Gene3D | G3DSA:3.30.730.10 | 2.6E-32 | 190 | 250 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 24.316 | 191 | 249 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 4.3E-35 | 191 | 255 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 7.85E-23 | 191 | 250 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 1.5E-14 | 191 | 241 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 9.2E-11 | 192 | 203 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 9.2E-11 | 215 | 231 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 302 aa Download sequence Send to blast |
MALDHVSALE LIKHHLLGEF SPVKSCATEP SDPGSSFTQN QWSPEDNSDL SRSQSESFCS 60 KSSSTFDSSI EISDYLDSSD IFEFNSAGFF DFEEKPQAID LTTPKSTNLS SQNSFEFDSK 120 PLVSDDYFEF EQKPQILKQT DPKPANSIRK PALTISLPNK TEWIQFASSD QAAVKPATVQ 180 KASSNEEKKK HYRGVRQRPW GKYAAEIRDP NRRGSRVWLG TFDTAIEAAR AYDRAAFKLR 240 GAKSILNFPL EAGKAEPVAS VERKRRRQEH REDAKAVVAV KKEKLTETPL TPSSWMAFWD 300 S* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2gcc_A | 1e-28 | 188 | 256 | 2 | 70 | ATERF1 |
| 3gcc_A | 1e-28 | 188 | 256 | 2 | 70 | ATERF1 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 262 | 266 | RKRRR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. Involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways. {ECO:0000269|PubMed:10715325, ECO:0000269|PubMed:9756931}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00549 | DAP | Transfer from AT5G47230 | Download |
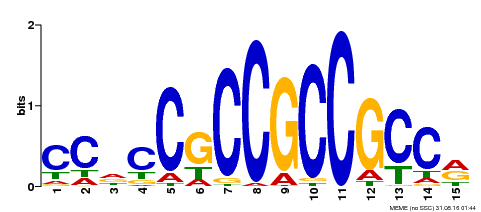
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pbr035774.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Ethylene induction is completely dependent on a functional ETHYLENE-INSENSITIVE2 (EIN2). Wounding as well as cold stress induction does not require EIN2. Transcripts accumulate strongly in cycloheximide-treated plants, a protein synthesis inhibitor. Seems to not be influenced by jasmonate, Alternaria brassicicola, exogenous abscisic acid (ABA), cold, heat, NaCl or drought stress. {ECO:0000269|PubMed:10715325}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | GU732432 | 0.0 | GU732432.1 Malus x domestica AP2 domain class transcription factor (AP2D8) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018505970.1 | 0.0 | PREDICTED: ethylene-responsive transcription factor 5-like | ||||
| Swissprot | O80341 | 3e-62 | EF102_ARATH; Ethylene-responsive transcription factor 5 | ||||
| TrEMBL | A0A498JN73 | 0.0 | A0A498JN73_MALDO; Uncharacterized protein | ||||
| STRING | XP_008355567.1 | 1e-160 | (Malus domestica) | ||||
| STRING | XP_008369419.1 | 1e-160 | (Malus domestica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1085 | 34 | 109 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G47230.1 | 2e-53 | ethylene responsive element binding factor 5 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



