 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pbr036239.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Maleae; Pyrus
|
||||||||
| Family | ARR-B | ||||||||
| Protein Properties | Length: 638aa MW: 71261.1 Da PI: 6.4222 | ||||||||
| Description | ARR-B family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 75.9 | 5.2e-24 | 199 | 252 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
k+r++W+ +LH++Fv+av+q+ G +k Pk+il+lm+v+ Lt+e+v+SHLQkYRl
Pbr036239.1 199 KARVVWSVDLHQKFVKAVHQI-GFDKVGPKKILDLMNVPWLTRENVASHLQKYRL 252
68*******************.9*******************************8 PP
| |||||||
| 2 | Response_reg | 80.7 | 4.8e-27 | 19 | 127 | 1 | 109 |
EEEESSSHHHHHHHHHHHHHTTCEEEEEESSHHHHHHHHHHHH..ESEEEEESSCTTSEHHHHHHHHHHHTTTSEEEEEESTTTHHHHHHHHHTTES CS
Response_reg 1 vlivdDeplvrellrqalekegyeevaeaddgeealellkekd..pDlillDiempgmdGlellkeireeepklpiivvtahgeeedalealkaGak 95
vl+vdD+p+ +++l+++l+k y ev+++ ++eal ll+ek+ +D+++ D++mp+mdG++ll++ e +lp+i+++ ge + + + ++ Ga
Pbr036239.1 19 VLVVDDDPTWLKILEKMLKKCSY-EVTTCGLAREALCLLREKKngYDIVISDVNMPDMDGFKLLEHVGLEM-DLPVIMMSVDGETSRVMKGVQHGAC 113
89*********************.***************999999**********************6644.8************************ PP
EEEESS--HHHHHH CS
Response_reg 96 dflsKpfdpeelvk 109
d+l Kp+ ++el++
Pbr036239.1 114 DYLLKPIRMKELRN 127
***********987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PIRSF | PIRSF036392 | 3.7E-149 | 1 | 595 | IPR017053 | Response regulator B-type, plant |
| SuperFamily | SSF52172 | 8.25E-35 | 16 | 140 | IPR011006 | CheY-like superfamily |
| Gene3D | G3DSA:3.40.50.2300 | 9.8E-44 | 16 | 146 | No hit | No description |
| SMART | SM00448 | 2.3E-32 | 17 | 129 | IPR001789 | Signal transduction response regulator, receiver domain |
| PROSITE profile | PS50110 | 42.452 | 18 | 133 | IPR001789 | Signal transduction response regulator, receiver domain |
| Pfam | PF00072 | 4.9E-24 | 19 | 128 | IPR001789 | Signal transduction response regulator, receiver domain |
| CDD | cd00156 | 2.52E-29 | 20 | 133 | No hit | No description |
| SuperFamily | SSF46689 | 1.74E-16 | 197 | 255 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 4.0E-27 | 197 | 256 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 2.4E-21 | 199 | 252 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 6.9E-8 | 201 | 251 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000160 | Biological Process | phosphorelay signal transduction system | ||||
| GO:0009735 | Biological Process | response to cytokinin | ||||
| GO:0010082 | Biological Process | regulation of root meristem growth | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 638 aa Download sequence Send to blast |
MDSSFSSPRN DSFPAGLRVL VVDDDPTWLK ILEKMLKKCS YEVTTCGLAR EALCLLREKK 60 NGYDIVISDV NMPDMDGFKL LEHVGLEMDL PVIMMSVDGE TSRVMKGVQH GACDYLLKPI 120 RMKELRNIWQ HVFRKKIHEI KDIESHESVE GIQLMRSGSD QYDEGYFLGA DDLTSSRKRK 180 DVDNKYDDKD FADCSSTKKA RVVWSVDLHQ KFVKAVHQIG FDKVGPKKIL DLMNVPWLTR 240 ENVASHLQKY RLYLGRLQKE NDLKSCFGGM KHSDSPSKDA QGSFGLQNSI SIQQNDFAHG 300 SFRVSGNNLV AQCVDPKSHE GDAKGIVSEP VAEIKKGSNG NTPDSQRTRS LQMVDFNNSC 360 VEPKNGVIGN VPDSQKIRSS PLDFNNSYAE TKKVLNGNIP DSQKIRTPQV RPSHSFTSVE 420 SEVNFTEFES TIRTKYSWNE IQLKKEQKPL IQLNSGFNKP PLPGPHSHFQ GDRLQSIPSI 480 SPRPSIAEGD VTGPAKSKPS YSEYINNQGS HVSPTISTAD SFPDQIKSCV VNHQVSESIS 540 TSTSNMKNQG FNLSSITDLE SAQRNLIVGS ASPFASLDDD FQICWFQGDC YGMNLGLQNI 600 EFPEYNDPAL ISEVPAHLYD ALRFDYPCDV THQNILS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 3e-17 | 195 | 257 | 1 | 63 | ARR10-B |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00010 | PBM | Transfer from AT1G67710 | Download |
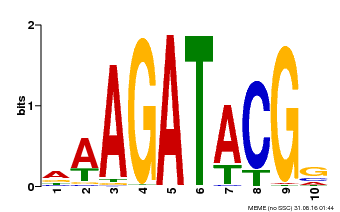
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pbr036239.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009359281.1 | 0.0 | PREDICTED: two-component response regulator ARR11 isoform X1 | ||||
| TrEMBL | A0A498HPI3 | 0.0 | A0A498HPI3_MALDO; Two-component response regulator | ||||
| STRING | XP_009359281.1 | 0.0 | (Pyrus x bretschneideri) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3999 | 33 | 60 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G67710.1 | 1e-128 | response regulator 11 | ||||



